Back to Journals » Infection and Drug Resistance » Volume 14
Bacterial Pathogens, Drug-Resistance Profile and Its Associated Factors from Patients with Suspected Peritonitis in Southern Ethiopia
Authors Alelign D , Ameya G, Siraj M
Received 19 August 2021
Accepted for publication 25 September 2021
Published 5 October 2021 Volume 2021:14 Pages 4107—4117
DOI https://doi.org/10.2147/IDR.S335103
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Dagninet Alelign,1 Gemechu Ameya,2 Munira Siraj1
1Department of Medical Laboratory Sciences, College of Medicine and Health Sciences, Arba Minch University, Arba Minch, Ethiopia; 2Department of Medical Laboratory Sciences, College of Medicine and Health Sciences, Kotebe Metropolitan University, Addis Ababa, Ethiopia
Correspondence: Dagninet Alelign Tel +251 931-155-573
Email [email protected]
Background: Ascitic fluid plays a critical role in the microbiological diagnosis of peritonitis. Drug-resistant bacterial infection of the peritoneal cavity is becoming a public health threat. However, data on bacterial profile and antimicrobial-resistant pattern of isolates from the ascitic fluid are scarce. Thus, this study was aimed to assess drug-resistant bacteriological profiles and factors associated with peritonitis in southern Ethiopia.
Methods: An institutional-based cross-sectional study was conducted from March 2019 to December 2019. A semi-structured questionnaire was used to collect socio-demographic and clinical data. A total of 147 ascitic fluid samples were aseptically collected and inoculated onto blood agar, MacConkey agar, and chocolate agar. The inoculated culture media were incubated aerobically and micro-aerobically at 37°C for 48 hrs. Bacterial identification was done by standard protocols and the antimicrobial susceptibility testing by Kirby Bauer’s disk diffusion method. Logistic regression was used to identify the associated factors with bacterial peritonitis.
Results: Of the total study participants, the overall magnitude of bacterial peritonitis was 19.05% with a total of 30 bacterial isolates. Majority of the isolates were Gram negative bacteria with predominant species E. coli 36.67% followed by Gram positive S. aureus 13.33%. The multidrug resistant isolates accounts about 43.3% while a quarter of isolated S. aureus were methicillin resistant. The bacterial peritonitis was associated with recent history of surgery [AOR = 8.724, 95% CI: (2.688– 28.314)], hospitalization more than seven days [AOR = 8.990, 95% CI: (2.755– 29.342)], cirrhosis [AOR = 2.751, 95% CI: (1.109– 6.822)] and alcoholism [AOR = 5.802, 95% CI: (1.948– 17.285)].
Conclusion: Nearly half of the isolated bacteria were observed to be MDR, and this may alarm all healthcare workers and policymakers. Thus, continuous surveillance of antimicrobial resistance patterns along with associated factors is essential for regular monitoring of transmission of drug-resistant bacteria and the emergence of antibiotic resistance.
Keywords: antimicrobial resistance, ascitic fluid, bacteriological profile, peritoneal effusion, southern Ethiopia
Introduction
Body fluids are important in transporting nutrients as well as waste products, regulating body temperature, and assessing the respiration process. Naturally, body fluids are sterile under normal circumstances and the presence microorganisms indicates the infection.1–3 Different infectious conditions could change the physicochemical nature of the fluids. Typically, bacterial infections have greater clinical urgency. Even a single colony of potentially pathogenic bacteria can be significant to cause infection.4,5
Bacterial infections of the peritoneal site have been currently substantial clinical challenges that could be life-threatening.6,7 Translocation of gut bacteria and their product is the major clinical source of peritoneal infections by reduction of intestinal motility, alteration of the gut’s barrier function, and local immune responses.8,9 Though it’s rare, peritonitis can develop without an apparent intra-abdominal source of infection, called spontaneous bacterial peritonitis (SBP).10,11 Abdominal surgical procedures, trauma, rupture of the appendix, bowel obstruction, pancreatitis, chronic diseases (HIV/AIDS, hepatitis, cirrhosis, and diabetes mellitus), and alcoholism may also enhance peritoneal site bacterial infection.10–13
Peritoneal effusion associated with bacterial infection is found to be in the range of 10% to 30%.6 About 40–70% of cases of bacterial peritonitis are associated with underlying conditions of cirrhosis, peritoneal dialysis, and post-surgical complications.14,15 The death of cirrhosis and peritoneal dialysis patients, with a mortality rate ranging from 18% to 40%, is the result of peritonitis coupled with the results of multiple organ failure.14,17 Around 60% of cirrhosis patients develop bacterial peritonitis with an incidence rate of 7% to 30% per year.13,14 Nosocomial associated peritonitis is expected to be in the range of 25–30%, of which 15% of the infection rate is accounted by post-operative procedures.8,15
Clinically patients with peritonitis are characterized by abdomen distension with tenderness, fever, nausea, and vomiting, loss of appetite, inability to pass stool, and fatigue.14,18 The degree of severity of patients presenting with bacterial peritonitis could be affected by bacterial species and their products, antibiotic response, the intensity of concurrent infection, alteration of membrane permeability, and immunity status of the patient.8,19
Nowadays, the increasing relevance of multidrug-resistant bacterial infection challenges treatment success and putting people at increased risk for severe morbidity and subjected to recurrent hospitalization.9,20 Antimicrobial resistance profiles of bacteria depend on local antimicrobial prescribing practices and the prevalence of resistant strains of bacteria. The burden is expected to be high in a developing country where conventional treatment guidelines are practiced which contributes to the emergence of antibiotic resistance and increased patient population intrinsically at risk for nosocomial infections due to long-term hospitalization.19–21
In Ethiopia, there are limited studies about drug-resistant bacterial profiles from ascitic fluids.3,19,20 While identification of antimicrobial susceptibility pattern of bacteria is, critical for the better management and to combat the emergence of resistance and framing the antibiotic policy. Hence, this study was undertaken to determine antimicrobial-resistant patterns of isolated bacteria from ascitic fluids along with factors associated with bacterial peritonitis from southern Ethiopia.
Materials and Methods
Study Design and Setting
The institutional-based cross-sectional study design was conducted from March 2019 to December 2019. The study was conducted on inpatients of Arba Minch General Hospital, Southern Ethiopia. Arba Minch is located in the south part of Ethiopia at 454 km away from Addis Ababa the capital city of the country. The hospital provides preventive, curative, and rehabilitative care for more than 1.5 million people.
Study Population
Patients attending the inpatient ward of Arba Minch General Hospital were the source population of the study. While patients who have had peritoneal effusion with clinically suspected bacterial infection during the study period were enrolled as a study population.
Eligibility Criteria
All patients who had peritoneal effusion at inpatient wards of Arba Minch General Hospital during the study period were included. Severely ill patients unable to respond to questionnaires and ascitic fluids laid in rejection criteria (samples expected to be contaminated, incorrectly labeled, and delayed ascitic fluids for more than 2 hrs.) were excluded.
Sample Size Determination and Sampling Procedure
A total of 147 study participants, irrespective of age and sex, were recruited using the purposive sampling technique.
Data Collection Tool and Collection Processing
Data about socio-demographic characteristics, clinical and behavioral factors of the patient were collected by face-to-face administered semi-structured questionnaire. A pre-test was done to validate the questionnaire and required modification was done accordingly. Other important medical-surgical data of the patient were obtained from the patient medical record.
Specimen Collection and Processing
From eligible participants, about 2–3 mL of ascitic fluid was collected by the physician with a sterile test tube. The physical appearance of the fluids was recorded immediately after collection. A fluid with a clear appearance was centrifuged for 15 min at 2500–3000 rpm to concentrate the existing microbes; while the fluid with a turbid appearance was directly processed for microbiological analysis.19 All fluid samples were processed in a microbiology laboratory within 30 minutes after collection, if not processed within 30 minutes the sample was stored at 37°C incubator with nutrient broth medium.
Microbiological Isolation and Identification
The fluid samples were primarily inoculated into blood agar (Hi-Media Pvt. Ltd), chocolate agar (Hi-Media Pvt. Ltd), and MacConkey agar (Hi-Media. Ltd) within an aseptic procedure. The inoculated media were incubated at 37°C for 48hrs. Chocolate agar plate was incubated inside candle jar to provide 5–10% CO2 concentration for fastidious microaerophilic bacteria. After the incubation period plates were examined and any growth on the agar media was processed for identification and drug sensitivity test. Identification was done by colony characterization based on the physical appearance of colonies (shape, surface, margin, translucency, and pigment), hemolysis, lactose fermentation, Gram reaction, and finally selected biochemical tests were used for identification of the species of isolates as per the standard specified in Clinical and Laboratory Standards Institute (CLSI).22
Antimicrobial Susceptibility Testing
An antimicrobial susceptibility test was performed on Mueller Hinton agar by using Kirby-Bauer disk diffusion techniques.23 The inoculation was incubated at 35–37°C for 16–18 hours according to the CLSI protocols.22 Morphologically identical 3–5 pure colonies of bacteria on primary agar medium were suspended in nutrient broth with a reference to 0.5 McFarland standards. Then, by using a sterile swab the suspension was uniformly inoculated into Mueller Hinton agar medium (Oxoid. England). After 3–5 minutes of inoculation, selected antibiotic discs (Abtek company of UK) were aseptically placed on the surface of the medium and the diameter of the zone of inhibition was measured by millimeter after overnight incubation and interpreted as sensitive, intermediate, and resistant as per as the CLSI guidelines.22
The antibiotic disc was also selected according to CLSI guidelines.22 For Gram positive bacterial isolates; Penicillin (10 μg), Gentamicin (10 μg), Ciprofloxacin (5 μg), Tetracycline (30 μg), Doxycycline (30 μg), Clindamycin (10 μg), Erythromycin (15μg), Vancomycin (30μg), Cotrimoxazole (25 μg), Chloramphenicol (30 μg), Cefoxitin (30 μg), Cefepime (30 μg), and Ceftriaxone (30 μg) antibiotics were used. While for Gram negative bacterial isolates; Ampicillin (10 μg), Piperacillin (100 μg), Cefoxitin (30 μg), Cefepime (30μg), Ceftriaxone (5 μg), Cefuroxime (5 μg), Streptomycin (10 μg), Gentamicin (10 μg), Ciprofloxacin (5 μg), Tetracycline (30 μg), Doxycycline (30 μg), Meropenem (10 μg), Chloramphenicol (30 μg) and Ceftazidime (30 μg) antibiotics were used.
Data Quality Control
A pre-test was done on 5% of the sample size before the actual data collection time and the data collection tool was modified accordingly. In the laboratory analysis, standard operating procedures were strictly followed and laboratory materials and reagents were maintained inappropriate storage conditions along with quality control parameters. Quality control (QC) was performed to check the quality of culture media, biochemical test and antibiotic disk with standard control strains Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923, and Enterococcus faecalis ATCC 29212 based on the manufacturer’s instructions for QC test recommendations.22
Statistical Analysis
Data were edited, cleaned, and analyzed by SPSS version 25. Descriptive statistics like frequency, mean, and percentage were calculated to describe the demographic characteristics of the study population. Bivariate logistic regressions analysis was used to assess associations between bacterial peritonitis and potential associated factors. Odds ratio (OR) and 95% confidence interval (CI) were estimated. Variables with P <0.25 in the bivariable analysis were jointly entered into a multivariable analysis. The presence of associations and statistically significant was determined at a 95% confidence interval and P-value < 0.05.
Ethical Considerations
This study was ethically approved by the Institutional Review Board (IRB) of Arba Minch University College of Medicine and Health Sciences. Permission was obtained from Arba Minch General Hospital. Participation in the study was on a voluntary basis. All the study participants were informed about the purpose of the study and their rights to withdraw and the confidentiality of the obtained information. Informed verbal and written consent was obtained before the interview, and informed verbal and written assent was obtained from parents or guardians for assuring participants less than 18-year-old. Significant laboratory results were reported to the respective patients’ physicians for treatment. In general, this study was conducted under the Declaration of Helsinki.
Results
Socio-Demographic Characteristics of the Study Participants
A total of 147 patients having peritoneal effusion have participated in the study. The majority of the study participants (61.9%) were males and the age range of the study participant was from 13 to 82-year-old with a mean age of 51.06 + 15.4-year-old. Among age distribution, nearly half of the study participants were in the age group from 46 to 65-year-old. About 60.5% of the study participants were living in rural areas and nearly the same proportion attended a formal education [Table 1].
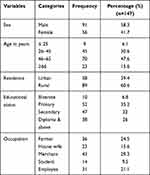 |
Table 1 Socio-Demographic Characteristics of Study Participants |
Clinical and Behavioural Characteristics of Study Participants
Among a total of 147 study participants, 11.5% of them had a recent history of abdominal surgical management for different clinical cases. The majority of the study participants’ body mass index was in the normal range (18.5–24.9 Kg/m2), and about 29.3% of study participants have had the habit of drinking alcohol more than three times per week. About 44% of study participants had cirrhosis.
Bacteriological Profile of Ascitic Fluids
Overall, 19.05% of ascitic fluids had bacterial growth with 95% CI (12.9–25.2). One fluid sample had mixed growth that makes the total number of isolated bacteria to be thirty. Nearly three-fourths (76.6%) of the isolated pathogen were Gram-negative. The frequently isolated bacteria were Escherichia coli 36.67% (11/30), followed by Klebsiella spp 20% (6/30), Staphylococcus aureus 13.33% (4/30), and Pseudomonas aeruginosa 13.33% (4/30) [Figure 1]. About 53.6% (15/28) half of the culture-positive ascitic fluids were observed among female participants. However, there was no statistically significant difference between gender [AOR=1.583, 95% CI=0.530–4.727, P value=0.411)] [Table 2].
 |
Table 2 Bivariate and Multivariable Logistic Regressions of Factors Associated to Bacterial Peritonitis |
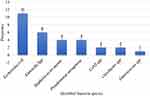 |
Figure 1 Distribution of isolated bacteria from ascitic fluids of study participant. |
Antibiotic-Resistant Pattern of Isolated Bacteria from Ascitic Fluids
Antibiotic-Resistant Pattern of Gram-Negative Bacteria Isolates
More than half of Gram-negative bacilli bacterial isolates were resistant to ampicillin, ciprofloxacin, cefepime, ceftriaxone, and cefuroxime, while gentamicin (69.6%) and meropenem (65.2%) showed better responses. The predominant isolate E. coli showed high resistance to ampicillin (72.7%), ceftriaxone (81.8%), and ciprofloxacin (81.8%). About 75% of P. aeruginosa isolates were resistance for cefepime and ciprofloxacin, while 75% of P. aeruginosa isolates were sensitive for piperacillin and meropenem [Table 3].
 |
Table 3 Antimicrobial Resistant Pattern of Gram Negative Bacteria Isolates from Ascitic Fluids |
Antibiotic-Resistant Pattern of Gram-Positive Bacterial Isolates
Among isolated Gram-positive bacteria, resistance was observed against penicillin (71.4%), tetracycline (71.4%), chloramphenicol (57.1%), and doxycycline (57.1%), while 57.1% to 71.4% of isolated Gram-positive bacteria were sensitive to cefoxitin, gentamicin, clindamycin, erythromycin, and cotrimoxazole. Out of S. aureus isolates 25% were methicillin resistance (MRSA). Similarly, among two coagulases negative Staphylococcus spp isolates one was methicillin resistance (MR-CoNS), whereas isolated Enterococcus spp (100%) were resistant to Vancomycin [Table 4].
 |
Table 4 Antimicrobial Resistant Pattern of Gram Positive Bacteria Isolates from Ascitic Fluids |
Multi-Drug Resistance (MDR) Bacterial Isolates from Ascitic Fluids
Out of 30 isolated pathogenic bacteria, 43.3% of them were found to have multidrug resistance (MDR). Multidrug resistance Gram-negative and Gram-positive bacterial isolates were found to be 43.5% (10/23) and 42.9% (3/7), respectively. More than half of isolated E. coli and Enterobacter spp were MDR [Table 5].
 |
Table 5 Multi-Drug Resistant Pattern of Isolated Bacteria from Ascitic Fluids |
Associated Factors for Bacterial Peritonitis (Culture-Positive Ascitic Fluids)
Both bivariable and multivariable analysis models of logistic regressions were done to determine factors associated with bacterial infection of the peritoneal cavity. The variables with a p-value less than or equal to 0.25 in the bivariable analysis were collectively enrolled to multivariable logistic regression analysis for the final identification of factors independently associated with the bacterial peritonitis. Among hypothesized factors, patients who had undergone abdominal surgery were 8.72 times more likely to develop peritoneal cavity bacterial infection [AOR= 8.724, 95% CI: (2.688–28.314)]. Likewise, hospitalized patients for more than seven days were 8.99 times more likely to develop bacterial infections of the peritoneal cavity [AOR= 8.990, 95% CI: (2.755–29.342)]. Patients presented with cirrhosis were 2.75 times more likely to develop peritoneal cavity bacterial infection [AOR= 2.751, 95% CI: (1.109–6.822)]. And patients who had a habit of alcohol drinking were 5.8 times at risk to develop bacterial infections of the peritoneal cavity [AOR= 5.802, 95% CI: (1.948–17.285)] [Table 2].
Discussion
The peritoneal cavity is a sterile site in which no bacteria or any microbes exist as commensals in a healthy state. Any microbe isolated from this site is considered a significant pathogen. The pathogenesis of peritoneal infection is determined by bacterial factors which influence the transition from contamination to infection. Bacteria responsible for peritoneal fluid infection may vary due to different socio-demographic, clinical or medical, and behavioral factors of personnel.3,5,20
In this study, the overall bacterial infection rates of peritoneal cavities were 19.04% [95% CI (12.9, 25.2)]. This finding was in line with studies conducted in North India (15.3%),5 Bangladesh (22.8%),24 Hubli, India (25.3%),3 and Nile Delta (25.7%).25 In contrast, our finding was highly lower than studies done in New Delhi, India (35.1%),4 Kanpur, India (42.8%),26 Mumbai, India (58%),11 Korea (78.6%)27 and Sudan (91.6%).16 However, our finding was greater than the study conducted in Addis Ababa Ethiopia (5.8%)19 and Rama, Kanpur (11.8%).2 The reason for the discrepancy of the result might be due to the emergence of peritoneal effusion associated with different underlying diseases and implementation difference of infection prevention programs as well as the number of study population differences.
Gram-negative bacteria were predominantly isolated (76.67%), accounted by E. coli 36.67%, Klebsiella spp 20%, and P. aeruginosa 13.33%, while isolated Gram-positive bacteria constitute only 23.33% with the maximum number of S. aureus 13.33%. This was in line with studies conducted in Kanpur, India,26 Delhi, India,4 and Northern, India.5 However, a retrospective study conducted among cirrhotic and peritoneal dialysis patients in Nile Delta, Egypt,25 Sudan,16 and Maryland, USA6 showed that Gram-positive bacteria were predominantly isolated. The observed difference might be due to study population difference and pre-sampling performed medical procedures since types of bacteria depend on where it was translocated.
The antimicrobial susceptibility pattern of this study showed that more than 56.5% of Gram-negative bacterial isolates were resistant to ampicillin (56.5%), cefuroxime (56.5%), cefepime (65.2%), ciprofloxacin (69.6%), and ceftriaxone (69.6%). This implies that those commonly prescribed antibiotics will have a probability of less than fifty percent to be used as empirical therapy for bacterial peritonitis particularly in the study area. However, isolated Gram-negative bacteria showed relatively good sensitivity to gentamicin (69.6%) and meropenem (65.2%). About 75% of P. aeruginosa isolates were sensitive for piperacillin and gentamicin. The finding of this study was comparable to previous studies conducted in Karnataka, India,3 Delhi, India,4 and Egypt.25 However, studies conducted from different sterile body fluids in Addis Ababa and Mekelle, Ethiopia19,20 Gram-negative bacterial isolates were resistant to chloramphenicol and gentamicin. The reason for this discrepancy could be due to the practice of antibiotic prescription and the distribution difference of resistant bacteria.
Gram-positive bacterial isolates were also showed resistance to penicillin (71.4%), tetracycline (71.4%) and doxycycline (57.1%), and chloramphenicol (57.1%), while 57.1% to 71.4% of isolated Gram-positive bacteria were sensitive to cefoxitin, gentamicin, clindamycin, erythromycin, and cotrimoxazole. 25% of isolated S. aureus were methicillin resistance (MRSA) while 50% of isolated coagulase-negative Staphylococcus spp were methicillin resistance (MR-CoNS). Likewise, isolated Enterococcus spp were resistant to vancomycin. The finding was comparable to the studies conducted in Ballari, India,2 and Kanpur, India.26 In contrast studies conducted in Karnataka, India,3 Addis Ababa, Ethiopia,19 and Mekelle, Ethiopia20 reported that isolated Gram-positive bacteria had higher resistance to erythromycin, clindamycin, and cotrimoxazole. This might be due to trends in the prescription practice of antibiotics, the strain of isolates, and the distribution of drug-resistant bacteria.
Overall, multidrug resistance (MDR) bacterial isolates were 43.3% (13/30) among 28 culture-positive peritoneal fluids. MDR Gram-negative and Gram-positive bacterial isolates were found to be 43.5% (10/23) and 42.9% (3/7), respectively, which calls for the immediate attention of healthcare providers and policymakers for prudent antibiotic use in the study area. The result was comparable to studies conducted in Delhi, India,4 and Northern India.5 However, the total level of MDR isolates in this study was lower than studies conducted from various body fluids at Addis Ababa and Mekelle, Ethiopia.19,20 This may be due to frequent prescription of broad-spectrum antibiotics for empirical therapy, distribution of drug-resistant bacteria, and isolated strain difference.
Variables that may have contributed to the pathophysiology of ascitic fluids associated with bacterial peritonitis were analyzed. Among these, the recent history of abdominal surgery was one of the significantly associated factors among culture-positive ascitic fluids. This was similar to independent studies conducted in Boston, USA,8 and Cincinnati, USA.9 The reason might be due to translocation of skin and gut normal flora bacteria and their products into the sterile cavities of the peritoneal during the procedures. Bacteria may also be introduced via instrumentations unless it is fully sterilized. Despite this, prolonged postoperative length of stay of the patient in a hospital may also lead to recurrent infection.
Another important factor in this study was hospitalization (stay of patients in the hospital for more than seven days) showed a statistically significant association with bacterial peritonitis. Similarly, different studies in Alexandra, UK,15 and Rome, Italy28 also narrated in associated with nosocomial infections in different aspects. In fact, long-term hospitalization leads to nosocomial infection, this might be due to the level of environmental hygiene of the hospital that can be used as a source of bacterial infection via mechanical ventilation or physical contact of patients with different fomites. Moreover, different medical and surgical invasive procedures which are taken as a treatment for other related health problems might introduce bacteria into the sterile sites and leads to infection. In addition, personnel might fail to follow the basic infection control procedures such as hand washing between patient contacts.
Another underlying clinical factor that had shown statistically significant association with bacterial peritonitis was patients with cirrhosis. This was similar to studies conducted in Sao Paulo, Brazil,13 Alexandra, UK,15 and Maryland, USA.6 This might be due to patients with cirrhosis leads to immune dysfunction as a result of low complement levels and protein synthesis, poor phagocytic activity, in parallel with a state of excessive activation of pro-inflammatory mediators (interleukin-1, interleukin-6, tumor necrosis factor-alpha). Thus, bacteria can easily pass into the ascitic fluid through the damaging membranes and leading to bacterial overgrowth and develops infection. Likewise, the changes in bile secretion created a more favorable environment for the survival of foreign bacteria in the intestinal tract. Even, oral symbiotic bacteria in liver cirrhosis patients can invade the intestine wall as a result of bile secretion changes.17
The patient who had a habit of alcohol drinking was also another behavioral factor that was statistically significant to bacterial infection of the peritoneal cavity. This result agrees with studies in Mumbai, India.11 The reason might be due to frequent alcohol consumption affects the contribution of the intestinal microbial populations by disturbing the balance of intestinal homeostasis. In addition, alcohol metabolite, acetaldehyde, has also a direct effect on gut epithelial cell function by disrupting tight junctions by accelerating inflammatory responses with a significant alteration of the immune system and may thus alter surround membrane permeability and enhance bacterial translocation from the intestines.29
Limitation of the Study
This study was limited by the fact that conventional culture methods were used for the identification of the organisms, as a result, most of the isolated bacterial species could not identify into strain level. Besides, the presence of strict anaerobic and fastidious bacteria might also affect the outcome of our result. The antimicrobial tests recommended using the minimum inhibitory concentration (MIC) method were not carried out because of budget constraints. The event of the use of antimicrobials prior to sampling might also affect the result since the study populations were inpatients.
Conclusion
In this study, magnitudes of bacterial peritonitis were comparable to other similar studies. Gram-negative bacteria (E. coli followed by Klebsiella spp.) were the predominant etiologies of peritonitis. The majority of bacterial isolates had higher rates of resistance to commonly prescribed antibiotics. Recent history of surgery, hospitalization for more than a week, alcoholism, and cirrhosis variables were independently associated with culture-positive ascitic fluids. Thus, along with strengthened hospital infection prevention strategies, treatment and management of bacterial peritonitis should be based on the knowledge of bacterial etiology and their antibiotic resistance patterns. Periodic monitoring of antimicrobial resistance patterns helps physicians to choose antimicrobial agents for empiric treatment of peritonitis.
Abbreviations
AOR, Adjusted Odd Ratio; ATCC, American Type Culture Collection; BMI, Body Mass Index; CoNS, Coagulase Negative Staphylococcus; COR, Crude Odd Ratio; MDR, Multi-Drug Resistance; MRSA, Methicillin Resistance Staphylococcus aureus; SBP, Spontaneous Bacterial Peritonitis.
Data Sharing Statement
The data sets analyzed during the current study are not publicly available due to ethical and confidentiality reasons but are available from the corresponding author on reasonable request under the Ethics Committee’s approval.
Acknowledgments
We would like to thank Arba Minch University, Colleges of Medicine and Health Sciences, Department of Medical Laboratory Science for giving us material and reagent support. We are very grateful to Arba Minch General Hospital staff for their cooperating during data collection. Finally, we would like to acknowledge study participants for their preparedness.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Balfe A, Barry S, Blake O, et al. The biochemistry of body fluids. In: McGing P, O'Kelly R, O'Meara Y, editors. ACBI Scientific Committee Guidelines.
2. Vishalakshi B, Hanumanthappa P, Krishna S. A study on aerobic bacteriological profile of sterile body fluids. Int J Curr Microbiol App Sci. 2016;5:120–126. doi:10.20546/ijcmas.2016.505.013
3. Harshika YK, Shobha MKR, Patil AB, Smita NR. A study on bacteriological profile and antimicrobial resistance pattern from various body fluids of patients attending the tertiary care Hospital, KIMS, Hubli. Indian J Microbiol Res. 2018;5(4):530–534.
4. Sharma R, Anuradha ND. Bacteriological profile and antimicrobial sensitivity pattern in sterile body fluids from a tertiary care hospital. J Appl Microbiol Biochem. 2017;1(1):1.
5. Rouf M, Nazir A. Aerobic bacteriological profile and antimicrobial sensitivity pattern of bacteria isolated from sterile body fluids: a study from a tertiary care hospital in North India. Microbiol Res J Int. 2019;21:1. doi:10.9734/mrji/2019/v28i130123
6. Hardick J, Won H, Jeng K, et al. Identification of bacterial pathogens in ascitic fluids from patients with suspected spontaneous bacterial peritonitis by use of road-based PCR (16S PCR) coupled with High-Resolution Melt Analysis (HRMA). J Clin Microbiol. 2012;9:JCM–00345.
7. Sofjan AK, Musgrove RJ, Beyda ND, et al. Prevalence and predictors of spontaneous bacterial peritonitis due to ceftriaxone-resistant organisms at a large tertiary centre in the USA. J Glob Antimicrob Resist. 2018;15:41–47. doi:10.1016/j.jgar.2018.05.015
8. Goldstein EJ, Snydman DR. Intra-abdominal infections: review of the bacteriology, antimicrobial susceptibility and the role of ertapenem in their therapy. J Antimicrob Chemother. 2004;53(suppl_2):29–36. doi:10.1093/jac/dkh201
9. Solomkin JS, Mazuski JE, Baron EJ, et al. Guidelines for the selection of anti-infective agents for complicated intra-abdominal infections. Clin Infect Dis. 2003;37(8):997–1005. doi:10.1086/378702
10. Barraclough K, Hawley CM, McDonald SP, et al. Polymicrobial peritonitis in peritoneal dialysis patients in Australia: predictors, treatment, and outcomes. Am J Kidney Dis. 2010;55(1):121–131. doi:10.1053/j.ajkd.2009.08.020
11. Bankar SS, De AS, Baveja SM. Study of ascitic fluid for diagnosis of spontaneous bacterial peritonitis (SBP) in adult patients with cirrhosis. Inter J Medic Appl Sci. 2014;3(1):41–49.
12. Soriano G, Castellote J, Álvarez C, et al. Secondary bacterial peritonitis in cirrhosis: a retrospective study of clinical and analytical characteristics, diagnosis and management. J Hematol. 2010;52(1):39–44.
13. Reginato TJ, Oliveira MJ, Moreira LC, Lamanna A, Acencio MM, Antonangelo L. Characteristics of ascitic fluid from patients with suspected spontaneous bacterial peritonitis in emergency units at a tertiary hospital. Sao Paulo Med J. 2011;129(5):315–319. doi:10.1590/S1516-31802011000500006
14. European Association for the study of the liver. EASL clinical practice guidelines on the management of ascites, spontaneous bacterial peritonitis, and hepato-renal syndrome in cirrhosis. J Hepatol. 2010;53(3):397–417. doi:10.1016/j.jhep.2010.05.004
15. Acevedo J. Multiresistant bacterial infections in liver cirrhosis: clinical impact and new empirical antibiotic treatment policies. World J Hepatol. 2015;7(7):916. doi:10.4254/wjh.v7.i7.916
16. Saint Paul LP, Albessard F, Gaillard C, et al. Improved conventional method for the laboratory diagnosis of peritonitis from peritoneal dialysates in Sudan. Dial Transplant. 2008;23:27–32.
17. Li PK, Szeto CC, Piraino B, et al. Peritoneal dialysis-related infections recommendations: 2010 update. Perit Dial Int. 2010;30(4):393–423. doi:10.3747/pdi.2010.00049
18. El-Bedewy TA, El-Sebaey MA, Okda HI, El-Deen MA. Microbiological study of spontaneous bacterial peritonitis in Tanta university hospitals: empirical antibiotic therapy. Egypt Liver J. 2017;7(1 and 2):5–8. doi:10.1097/01.ELX.0000525968.17584.d3
19. Teklehymanot F, Legese MH, Desta K. Bacterial profile and their antimicrobial resistance patterns from body fluids at Tikur Anbessa specialized hospital, Addis Ababa, Ethiopia. Biol Med. 2017;9(408):2. doi:10.4172/0974-8369.1000408
20. Tsegay E, Hailesilassie A, Hailekiros H, Niguse S, Saravanan M, Abdulkader M. Bacterial isolates and drug susceptibility pattern of sterile body fluids from tertiary hospital, Northern Ethiopia: a four-year retrospective study. J Pathog. 2019;2019:6. doi:10.1155/2019/5456067
21. Fiore M, Maraolo AE, Gentile I, et al. Current concepts and future strategies in the antimicrobial therapy of emerging gram-positive spontaneous bacterial peritonitis. World J Hepatol. 2017;9(30):1166. doi:10.4254/wjh.v9.i30.1166
22. Wayne PA; Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
23. Hudzicki J. Kirby-Bauer disk diffusion susceptibility test protocol. In: ASM Microbe Library. USA: American Society for Microbiology; 2009. Available from: http://www.microbelibrary.org/component/resource/laboratory-test/3189-kirby-bauer-diskdiffusion-susceptibility-test-protocol.
24. Sarker JA, Alam MS, Khan M, Mahtab M-A, Ashraf MS, Khondaker FA. Variant of ascitic fluid bacterial infections in patients of liver cirrhosis. Euroasian J Hepatogastroenterol. 2015;5(2):131. doi:10.5005/jp-journals-10018-1152
25. Khalil HS, Elkhalawany W, Elhendawy M, Badawi R, Abdelwahab MA, Abd-Elsalam S. Identification of ascitic fluid bacterial pathogens in spontaneous bacterial peritonitis in Nile Delta and its impact on clinical outcome of these patients. Microbiol Res J Int. 2016;1–6. doi:10.9734/BMRJ/2016/29869
26. Sujatha R, Pal N. Bacteriological profile and antibiotic sensitivity pattern from various body fluids of patients attending Rama Medical College Hospital, Kanpur. Int J Adv Case Rep. 2015;2(3):119–124.
27. Yoon SH, Choi NW, Yun SR. Detecting bacterial growth in continuous ambulatory peritoneal dialysis effluent using two culture methods. Korean J Intern Med. 2010;25(1):82. doi:10.3904/kjim.2010.25.1.82
28. Merli M, Lucidi C, Di Gregorio V, et al. The spread of multi drug resistant infections is leading to an increase in the empirical antibiotic treatment failure in cirrhosis: a prospective survey. PLoS One. 2015;10(5):e0127448. doi:10.1371/journal.pone.0127448
29. Tuomisto S, Pessi T, Collin P, Vuento R, Aittoniemi J, Karhunen PJ. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 2014;14(1):1–8. doi:10.1186/1471-230X-14-40
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
