Back to Journals » Nanotechnology, Science and Applications » Volume 14
Bacterial Biofilm Destruction: A Focused Review On The Recent Use of Phage-Based Strategies With Other Antibiofilm Agents
Authors Amankwah S , Abdella K, Kassa T
Received 19 June 2021
Accepted for publication 20 August 2021
Published 14 September 2021 Volume 2021:14 Pages 161—177
DOI https://doi.org/10.2147/NSA.S325594
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Israel (Rudi) Rubinstein
Stephen Amankwah,1,2 Kedir Abdella,1 Tesfaye Kassa1
1School of Medical Laboratory Sciences, Institute of Health, Jimma University, Jimma, Ethiopia; 2Accra Medical Centre, Accra, Ghana
Correspondence: Tesfaye Kassa
School of Medical Laboratory Sciences, Institute of Health, Jimma University, P. O. Box 788, Jimma, Ethiopia
Tel +251931057195
Email [email protected]
Abstract: Biofilms are bacterial communities that live in association with biotic or abiotic surfaces and enclosed in an extracellular polymeric substance. Their formation on both biotic and abiotic surfaces, including human tissue and medical device surfaces, pose a major threat causing chronic infections. In addition, current antibiotics and antiseptic agents have shown limited ability to completely remove biofilms. In this review, the authors provide an overview on the formation of bacterial biofilms and its characteristics, burden and evolution with phages. Moreover, the most recent possible use of phages and phage-derived enzymes to combat bacteria in biofilm structures is elucidated. From the emerging results, it can be concluded that despite successful use of phages and phage-derived products in destroying biofilms, they are mostly not adequate to eradicate all bacterial cells. Nevertheless, a combined therapy with the use of phages and/or phage-derived products with other antimicrobial agents including antibiotics, nanoparticles, and antimicrobial peptides may be effective approaches to remove biofilms from medical device surfaces and to treat their associated infections in humans.
Keywords: biofilms, extracellular polymeric substance, phages, phage-derived enzymes, nanoparticles, antimicrobial peptides
Introduction
Despite being found naturally as individual cells in planktonic form, most bacteria are capable of forming multiple cell structures called biofilms by adhering to biotic and abiotic surfaces. Bacterial biofilm formation is regarded as one of the most important strategies of survival that increases the virulence to be more pathogenic and consequently contributes to further resistance to antibiotics.1 Biofilms formed on human tissues and surfaces of medical devices, including implants, are implicated in the pathogenesis of chronic bacterial infections, such as urinary tract infections, pneumonia, orthopedic infections, oral infections, wound infections, and cystic fibrosis among others.2,3 Bacterial biofilms, despite their beneficial use for biological stages of wastewater treatment, waste reactors, numerous food production systems, and space travel,4 have enormous negative implications on human health.
Bacteriophages or phages, are viruses that infect bacterial cells with high specificity. They are the dominant biological entities in the planet and those phages with virulent or lytic to bacteria are also capable of infecting and destroying biofilm-forming bacteria.5 The threat posed by increasing incidence of antimicrobial-resistant (AMR) bacteria coupled with the paucity of new antibacterial drug intervention has revived interest in using bacteriophages against AMR. Similarly, the ineffectiveness of antibiotics on biofilms has resulted in a growing scientific interest in phages as an alternative strategy in controlling and preventing biofilm formation.6 Indeed, the interaction of phages and biofilms as a subject of research in scientific publications have risen exponentially in the last decade.2,7–9 Reports from the majority of studies on application of phages and phage-encoded proteins as alternative approaches to control and prevent biofilms formed particularly by ESKAPEE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp. and Escherichia coli) are encouraging.10–15
In the search for effective alternative approaches to combat biofilm-forming bacteria and their associated infections, some advanced and relevant techniques for biofilm destruction have been intensely emphasized. These strategies include biofilm degradation by electrochemical method which is a combinatorial effect of applying lower dose antibiotics in a weak electric field, the use of antimicrobial lipids as biofilm inhibitors and inhibition of alarmone scheme by natural peptides in response to environmental stress.16 Others include interruption of molecular system of biofilm formation and irrigation techniques including passive ultrasonic, subsonic, or laser-activated irrigation for removal of root canal biofilms.17,18 Among these techniques, the use of phage-based strategies was least emphasized, whereas they are economical and potentially modifiable. It is thus essential to discuss phages and bacterial biofilms. Therefore, the authors in this review present an overview of existing literature on bacterial biofilms and give an insight on the most recent use of phages as well as phage-derived enzymes as natural armament to destroy bacterial biofilms. The authors also illustrate the synergistic use of phages or phage-derived proteins and other antimicrobial agents that the former serve as vehicles for delivering antibiofilm agents to eradicate biofilms.
The Bacterial Biofilm
The Genesis of Bacterial Biofilms
Biofilm is a complex integrated aggregate of bacterial cells associated with adherence to both biotic and abiotic surfaces. This microbial or bacterial community is encased in a matrix of hydrated extracellular polymeric substances (EPS) produced predominantly by the cells themselves within the biofilm structure.1 The EPS matrix consists of polysaccharides (40–95%), proteins (1–60%), lipids (1–40%), nucleic acids (extracellular (eDNA) and RNA) (<1%), lipoproteins, enzymes, and inorganic components. Typically, 2–35% of the biofilm volume is composed of the microorganisms (less than 10% in bacterial biofilms) while the matrix accounts for about 90% of the total biofilm mass.19,20 Clearly, variation exists in the physical and chemical constituents of biofilm between different species of bacteria depending on the microorganisms’ type, stress level, nutrients availability, and host environment.21 As one of the critical steps in biofilm formation, the matrix provides structural support and protection for bacterial communities. Besides offering architectural stability and forming a defense shield from antimicrobial factors, the bacterial extracellular matrix plays alternative roles such as serving as signal targets and amplifiers, mediation of migration and colonization, capturing of cations, and exchange of genes.22 Pores and channels within the biofilm aid in the transport of nutrients, gases, water, and other molecules within the matrix and between the biofilm and the environment. It is noteworthy that the main component of the matrix is water (up to 97%), which baths the architectural and functional components of the matrix.1 In effect, by forming biofilm, bacteria are able to adapt to the surrounding environment and also able to survive in hostile environmental conditions.
Bacterial Biofilm Formation
Biofilm formation is a complex cooperative group process, which occurs in step-by-step processes and involves chemical communication within and between cells. This cell-to-cell communication system is coordinated by crosstalk of various signaling networks including two-component systems (TCS), diguanylate cyclase (DGC) systems, and quorum sensing (QS).2,23 The TCS, composed of histidine sensor kinase and response regulators, regulate signal transduction via phosphorylation or cyclic di-GMP (c-di-GMP), a secondary messenger, which permits DNA specific binding for modulation of gene expression. As a key signal network molecule synthesized by the DGC system, c-di-GMP level depends on membrane or cytoplasmic DGC and phosphodiesterase activities working solely or as part of TCS. Signal transduction by c-di-GMP involves allosteric modification of enzymes, interaction with transcription factor, riboswitch, and participation in post-transcriptional and post-translational processes within the cell. These activities by c-di-GMP coordinate the transition of bacteria from planktonic to biofilm growth mode.23
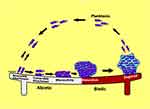
The QS system involves two main factors, the autoinducer (AI) and the receptor, which is based on population cell density or concentration. The AIs are small diffusible signaling molecules produced by the bacteria, which are detected by the receptors once a concentration threshold is achieved.24 Gram-negative and -positive bacteria secrete N-acyl homoserine lactones (AHL) and autoinducer oligopeptides (AIP) as signaling molecules, respectively, as well as AI-2.24,25 The binding of the AIs to their cognate receptors activates specific gene expression including biofilm formation and antibiotic resistance among others.23 Biofilm formation occurs within and between species of bacteria as well as interkingdom signaling among plants, fungi, and host cells, suggesting that microorganisms in a biofilm interact by self-talk, crosstalk and listen in.26 Biofilm formation proceeds in four stages: (i) adhesion (ii) microcolony formation (iii) maturation (iv) dispersion (Figure 1).
Adhesion
Bacterial attachment to a living or non-living surface begins the initial stage of biofilm formation with the generation of conditioning film which changes the physicochemical nature of the surface to initiate the adhesion process. Adhesion occurs through reversible and irreversible attachment processes.24 Reversible adhesion is a temporary attachment of the free-living bacteria to the conditioned surface, which causes a weak adhesion mediated by non-specific van der Waal’s, electrostatic and Lewis’s acid-based electronic forces.27 In contrast, irreversible adhesion is a permanent adhesion, which causes strong attachment of the bacteria to any surface, mediated by bacteria specific adhesion pili (fimbriae) and flagella. Bacterial adhesion is greatly affected and supported by EPS composition, nature of the surface, cell surface hydrophobicity, and flagella-pili-fimbriae movement. A rough surface provides a higher adhesion than a smooth surface. Likewise, due to the decreased in force of repulsion between the bacteria and the surface, hydrophobic and non-polar surfaces such as plastics and Teflon provide a higher adhesion than hydrophilic and polar surfaces such as metals and glasses.25,28
Microcolony Formation
Once adhesion is established, bacterial cells divide and multiply to develop three-dimensional (3D) clusters and aggregates called microcolonies. Microcolonies grow via cell proliferation to synthesize the EPS matrix for adhesion to surfaces, cohesion among cells, scaffolding cells together, maintaining the 3D architecture, and protection of the microcolonies. The EPS protects the microcolonies against various stresses such as mechanical removal, host immunity, metallic cations, oxidation, and antimicrobials with enhanced drug tolerance. EPS as a physical boundary can sequester or trap various substances to create nutritional and chemical gradient for the diffusion of oxygen, signaling molecules, inorganic ions, metabolites, and other solutes across the biofilm 3D architecture.29 Bacterial microcolonies normally consist of many types of micro-communities that coordinate with one another for substrates exchange, metabolic products dissemination, and metabolic end-products excretion. The EPS of Gram-positive bacteria is cationic and that of Gram-negative bacteria is neutral or polyanionic.25
Maturation
The formation of microcolonies from layered cells and small clusters leads to the generation of a thin biofilm to begin the maturation phase, with the synthesis of EPS matrix from main structural components (polysaccharide, protein, and eDNA). Clusters develop into macrocolonies (large microcolonies) with the displacement of cells from the substratum to form channels and voids, which facilitate the exchange of nutrients and waste products by infusing fluid into the biofilm.30 Polysaccharide forms the core of the matrix whereas eDNA is involved in horizontal gene transfer. The maturation of the biofilm through signal molecules, causes structural changes, as well as many changes in the expression of genes coding for different virulent factors. These changes include loss of cellular motility by expressing flagella-free phenotypes, reduction in protease and phospholipase C synthesis, decrease in the synthesis and release of toxins, and production of rough and sometimes mucus-like polysaccharide to better adapt to certain conditions of the biofilm microenvironment.23 The morphological changes in the biofilm enable metabolic adaptation under aerobic and anaerobic conditions that results in metabolically distinct microcolonies, whose presence may not only decrease or eliminate the time required to adapt to nutrients and oxygen stress but also provide an important metabolic context to resist antimicrobials present within the microenvironment.31
Dispersion
The final stage of biofilm formation is cell dispersion and the switch of sessile cells to the planktonic mode of growth, for colonization of new sites, to form a new cycle of biofilm. Dispersion is an active event induced or triggered by self-synthesized signaling molecules or cues such as fatty acid signaling molecules, and environmental conditions such as oxygen depletion, nutrient availability, starvation due to cessation of oxygen or nutrients, nitric oxide, and iron, which ultimately result in the overall reduction in the levels of c-di-GMP via series of post-transcriptional modifications.32 Low c-di-GMP levels, in turn, upregulate genes involved in cell motility, such as flagella synthesis or chemotaxis, and in EPS matrix degradation, such as matrix-degrading enzymes, endonucleases such as endA, which degrades DNA present in the matrix, and glycoside hydrolases such as pslG and pelA, which degrade the matrix polysaccharides Psl and Pel. Concurrently, genes involved in EPS production, such as polysaccharide synthesis, and in attachment, such as fimbriae synthesis, are downregulated. Phenotypes related with low c-di-GMP levels include increased motility, reduced adhesiveness, reduced matrix synthesis and dispersion.30,32
Burden of Bacterial Biofilm Occurrence in Medical Field
Biofilm control and prevention is an enormous problem currently for the food industry, agricultural sector and the medical field. The widespread occurrence of bacterial biofilms in every habitat on earth,33 including biofilms on medical device surfaces and in human tissues, pose a major threat causing chronic infections. According to the Center for Disease Control and Prevention (CDC), it is estimated that biofilms are responsible for over 65% of all chronic bacterial infections, while the National Institutes of Health (NIH) estimates around 80% of microbial infections and over 60% of nosocomial infections.32 Biofilm formation on or within medical devices, implants, and prostheses, cause device-associated infections such as urinary tract infections, orthopedic infections, endocarditis, periodontitis, gingivitis, osteomyelitis, cystic fibrosis, pneumonia, and wound infections, notably by multidrug-resistant (MDR) bacteria from the ESKAPEE group, as well as other Gram-negatives and -positives.34
Biofilm burden in medical field is exacerbated by antibiotics resistance of bacteria in biofilm communities contributing to persistent infections. With about 500–5000-fold increase in resistance to antibiotics compared with nomadic cells,35 bacterial biofilms formation has rendered conventional antibiotics ineffective and insufficient at eradicating biofilm-mediated infections.36 Furthermore, bacteria in biofilms are tolerant to antiseptic agents, germicides and the response of host immunity regardless of their location.24,36 The tolerance of bacterial biofilms to antimicrobials depends on several factors, which stem from different intrinsic and acquired resistance mechanisms of bacteria. Notable among them is the generation of semi-dormant cells from the deepest biofilm regions with reduced metabolic activity called persister cells.24 Biofilms contain a great population of persister cells, which are tolerant to all conventional therapeutics. As reported, resistance of bacteria in a biofilm may be due to (a) restriction of antibiotics diffusion by polymeric matrix, (b) interaction of the biofilm matrix with antibiotics that can retard and lower their activities, (c) action of the modifying enzymes such as β-lactamases or aminoglycoside adenylyltransferases, (d) alteration in metabolic activity inside the biofilm (chemical microenvironment), (e) genetic modifications of target cells or camouflaging the target sites, (f) slow growth rates of bacteria in which drugs are not effective, (g) generation of persister cells, which are tolerant to different antibiotics, (h) multiple microbial species, (i) extrusion of antibiotics using efflux pumps, and (j) the age of the biofilm. Thus, this multifactorial nature of bacterial biofilm formation and antimicrobial resistance impose great challenges for the adaptation of conventional antibiofilm therapeutic strategies.19,24
Bacteria–Phage Co-Interaction Within the Biofilm
Bacteriophages are considered to be the most abundant microorganisms on earth with numbers reported to be 10 times more than their bacterial hosts.37 Two types of phages exist depending on the life cycle: lytic or virulent phages, which exhibit lytic cycle by replicating their genome and subsequent release of assembled progeny phages causing rapid cell destruction and lysis of the host cells; and temperate or lysogenic phages, which exhibit lysogenic cycle by persisting as prophages within the genome of the host bacteria to achieve a co-existing state. The later cycle may undergo the lytic cycle following induction by environmental stimulus.38
As natural enemies of bacteria, phages are perfectly adapted to destruct biofilms using different mechanisms such as by degrading the extracellular matrix, penetrating the biofilm and infecting the bacteria (Figure 2). One of such mechanisms is the stimulation of the host bacteria to produce EPS-degrading enzymes. These host induced enzymes breakdown the rich polysaccharides and proteins within the extracellular matrix to facilitate phage penetration, replication, and elimination of the bacteria present in various metabolic states via lytic activity.5 In addition, phages can express enzymes with exopolysaccharide degrading activities (polysaccharide depolymerases) that degrade extracellular polymers by digesting the polysaccharide matrix and proteins in the biofilm that surrounds the bacteria as well as polysaccharide forming capsules and lipopolysaccharides. This process clears the bacterial protection barrier and then allows the entry of phage particles into the biofilm to replicate within the bacteria.27 High rates of phage replication occur given the high densities of bacteria in the biofilm structure. Phage induced bacterial lysis results in the release of progenies as they achieve local lysis of susceptible cells and as associated enzymes weaken the bacterial cell wall and degrade EPS within the biofilm.39 Lytic phages retain lytic activity against persister cells with reduced metabolic activity. Lysogenic phages can also integrate into the bacteria genome causing the bacteria to float naturally without adhering to surfaces to initiate the formation of mature biofilms.27
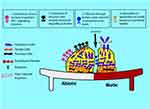 |
Figure 2 Schematic representation of phage mechanisms of biofilm destruction. |
Biofilms defensive mechanisms can resist phage infection by affecting phage adsorption, penetration, diffusion, and proliferation within the formed biofilms. Factors such as structure and thickness of the biofilm matrix, age of the biofilm, physiological heterogeneity within the biofilm, and the bacterial species or strains that form the biofilm in multispecies state, may limit phage infection and activity of the biofilm.40 Another interesting mechanism to prevent infection of phages is to specifically recognize the nucleic acids of the phages and destroy them. Bacteria use restriction-modification (R-M) system, defense island system associated with restriction modification (DISARM), prokaryote argonaute proteins (pAgos) and clustered regularly interspaced short palindromic repeats (CRISPR) – Cas9 to prevent phage infection.41 As a final barrier to phage infection, the bacteria can use an abortive infection system that leads to the death of the infected cell, preventing the spread of phages through the community.27
To overcome these resistance mechanisms, phages have developed several strategies, however. Phages are equipped with specific enzymes such as hydrolases, endolysins and depolymerases to overcome the structure, thickness, composition, and age of the biofilm with associated matrix as well as the bacterial cell structural parts.42 Phages can diffuse through biofilm water channels and penetrate the inner biofilm layers.43 Phages can also adsorb reversibly to the appendices of motile bacteria to penetrate inside the biofilm.44 Phages can tackle persister cells through the release of intracellular material which triggers the metabolism of the persister cells for phage infection and replication.2 Strategies by which phages escape the bacterial immune systems include the potential escape from R-M systems by lacking the endonuclease recognition site throughout the genome, acquisition of point mutations in the spacer sequence and production of anti-CRISPR protein which interferes with the system to escape the CRISPR/Cas9 system.45
The interaction of phages with bacteria is often seen as an antagonistic co-evolutionary cycle. The presence of phages may contribute to active biofilm formation as eDNA release through phage-mediated cell lysis by prophages, is responsible for horizontal gene transfer, which triggers stringent response of stabilizing the biofilm matrix.46,47 Phage interaction with bacteria may cause changes in the biofilm matrix leading to enhanced biofilm adhesion, virulence, dispersion of biofilms, colony variation and antibiotic tolerance.48 Owing to the co-evolution mechanism, phages are thus seen as actively involved in biofilm formation either as promoting or destructing agents.49
Application of Phages in Bacterial Biofilm Destruction
The tolerance of bacterial biofilms to antibiotics and host immunity has resulted in the search for alternative methods against bacteria in biofilms as well as antibiotic-resistant strains. Phages and phage-derived products have triggered scientists as the most important alternative to antibiotics in preventing and treating biofilms and associated infections. However, development of efficient phage-based treatments requires a deeper understanding of bacteria resistance to phages and co-evolutionary mechanisms between phages and bacteria, to minimize the likelihood emergence of resistance. Phage-based treatments for bacterial biofilm destruction includes the use of mono phages, phage cocktails, genetically engineered phages, and phage-derived enzymes.43 Some of the most recent application of phages and phage-derived products in bacterial biofilm destruction are summarized in this section (Figure 3).
 |
Figure 3 Phage-based treatment options in bacterial biofilm destructions. |
Application of Mono Phages
Application of mono phage in bacterial biofilm destruction involves the use of naturally occurring strictly virulent or lytic phages that do not encode genes for virulence, toxins or AMR. In addition, the phage should not be able to mediate horizontal gene transfer or transduce infected bacterial cells. Single phages usually have narrow host range as they are generally specific for a limited set of strains of the same bacterial species. Their applications as therapeutic and biocontrol agents in clinical, veterinary, environmental and food bacterial isolates are enormous and encouraging.50 Recently, phages PSTCR4 and PSTCR6, as part of 17 characterized novel phages, exhibited efficient reduction of well-established P. stuartii biofilms formed in catheter models. In addition, the phages showed killing capabilities in solid and liquid cultures in various patterns and levels of effectiveness. The phages, found to be free of virulence factors and resistance genes, suggest their potential as reliable therapeutic agents for phage therapy against P. stuartii biofilms in bacteriuric catheterized patients.51 In studying oral biofilm elimination as one of the main targets for caries prevention, phage SMHBZ8, isolated from human saliva samples, showed effective prevention of biofilm and reduction of existing biofilm of S. mutans in biofilm cultures and cariogenic dentin model.52 In a similar study, individual phages isolated from sewage samples prevented biofilm formation of S. mutans up to 97% by inhibiting the expression of genes involved in biofilm production.53 Both studies suggest that mono phages may be possible candidates of phage therapy for dental caries. To evaluate the application of phages in thermal and industrial environments, the effectiveness of single phages was assessed in biofilms of P. aeruginosa isolated from drinking and thermal water. The phages showed a maximum reduction of biofilms established on a stainless-steel coupon between treated and untreated surfaces.54 Interestingly, in a study on biofilms of methicillin resistant S. pseudintermedius, a zoonotic pathogen isolated from canine and veterinary workers, low concentrations of phages pSp-J and pSp-S applied, inhibited biofilm formation.55 Numerous mono phages have been applied to significantly inhibit or decrease to minimum levels of viable bacteria cells in biofilms and in liquid cultures with no considerable toxicity to mammalian cells. Notable among them are biofilms formed by E. faecalis,56 S. aureus57 and E. coli,58 all belonging to the ESKAPEE group. As revealed in these studies, anti-biofilm effects of lytic mono phages are concentration-dependent. Too high concentration of phages can interfere with the destruction of biofilm, and too low concentration of phages may not be adequate to infect and penetrate the biofilm. Therefore, treatment time is a major factor for bacterial biofilm destruction rather than the concentration of phages and so the appropriate concentration of phages should be applied according to the usage.55
Application of Phage Cocktails
Phage therapy is commonly applied in the form of phage mixture or cocktail targeting either mono or several bacterial strains. The logic behind the use of phage cocktails arises from the fact that simultaneous treatment targeting a variety of bacterial receptors with diverse antibacterial pathways will more efficiently decrease the bacterial burden, expand host range coverage, lysis potential, mitigate resistance or development of lysogenic strains.59 Simultaneous application of phages as found in several study models show greater efficacy in biofilm destruction than mono phage application.55,58
In a recent study, phage cocktail composed of four lytic phages, completely inhibited the growth of MDR E. coli and significantly prevented the development of biofilms compared with single phages. The phage mixture caused strong biomass reduction of biofilm and showed the highest biofilm inhibition up to nearly 87%.60 To broaden lytic spectra and increase the efficiency of therapeutic phage mixture, phages with and without polysaccharide degrading enzymes should be included in cocktails preparation. A study reported that a combination of 4 phages lysed all studied K. pneumoniae strains, although one of the phages lacked genes encoding polysaccharide depolymerases involved in degradation of the biofilm matrix.61 Similarly, Phages ΦKpnM-vB1, ΦKpnP-vB2 and ΦKpnM-vB3 isolated and characterized, did not only have lytic activity on K. pneumoniae and E. coli strains, but also, were highly efficient in reduction of Klebsiella biofilms when applied as a cocktail.62 In a Galleria mellonella infection model, co-incubation of monophage Sb-1 and PYO bacteriophage, two commercially available phage formulations, eradicated MRSA biofilm and prevented biofilm formation by completely abolishing heat production of MRSA.63 Unlike mono phage applications which are mostly lytic phages, phage cocktail could be a mixture of only lytic phages, temperate phages or both. Four temperate phages Trsa205, Trsa207, Trsa220 and Trsa222 in a cocktail, was capable of removing 65% of S. aureus biofilms, in addition to lysing two-thirds of the isolates.64 Due to their broad host range coverage, phage cocktails are effective on mixed-species biofilms. For example, phage cocktails AB-SA01 and AB-PA01 which target S. aureus and P. aeruginosa, respectively, when combined together, significantly reduced biofilm biomass in mixed-species biofilms, compared to the respective phage cocktail treatment.65 A mixture of two phages, philPLA-RODI and philPLA-C1C, showed a reduction in the amount of adhered bacterial cells to about 2 log units in both mono-species and dual-species biofilms of S. aureus and S. epidermidis.66 These studies suggest that application of phage cocktails other than individual phages in biofilm models, is highly efficient at destroying bacterial biofilms.
Application of Genetically Engineered Phages
Phages which lack enzyme-encoding genes are genetically engineered to express degradation enzymes for adsorption, penetration and diffusion of the phages through the EPS-matrix for biofilms destruction.67 For example, a modified T7DspB E. coli phage has been designed to express intracellularly a hydrolase that is released during infection as well as to the extracellular matrix enhancing biofilm degradation. Testing on E. coli biofilms showed the efficient expression of biofilm dispersing (DspB) enzyme during phage infection, causing biofilm degradation rate of about 99.997% and a decrease in the population of viable bacterial cells in the biofilm by 4.5 orders of magnitude, around 100 times higher than the efficiency of the parent T7 phage.68 Similarly, the construction of bacteriophage T4 Rnl1 exerts antibiofilm activity instead of bactericidal activity against S. mutans with a decrease in biofilm biomass resulting from scattered microcolonies and larger channels surrounded by minor or reduced exopolysaccharides.69 An engineered T7 bacteriophage encoded with a lactonase enzyme when constructed, expressed the AiiA lactonase to effectively degrade AHLS from many bacteria. Addition of the engineered phage to mixed-species biofilms of E. coli and P. aeruginosa inhibited biofilm formation.70 Some temperate phages with non-lytic features are genetically engineered into lytic phages with the production of endolysins useful for biofilm destruction and removal.71 With about half of the sequenced bacteria being lysogens, temperate phages could be easier to find than isolating lytic phages from nature.72
In a recent study, endolysin PM-477 of the type 1,4-beta-N-acetylmuramidase encoded on Gardnerella prophages when recombinantly expressed, demonstrated strong bactericidal activity against four different Gardnerella species. By domain shuffling, several engineered endolysins with 10-fold higher bactericidal activity than any wild-type enzyme was generated. When tested against a panel of 20 Gardnerella strains, the most active endolysin, PM-477, showed minimum inhibitory concentrations and had no effect on beneficial lactobacilli or other species of vaginal bacteria. Furthermore, when tested on vaginal samples of fifteen patients with either first time or recurring bacterial vaginosis, PM-477 killed the Gardnerella bacteria in thirteen of the cases and physically dissolved the biofilms without affecting the remaining vaginal microbiome. The high selectivity and effectiveness in eliminating Gardnerella, both in cultures of isolated strains as well as in clinically derived samples of natural polymicrobial biofilms, makes PM-477 a promising alternative to antibiotics for the treatment of bacterial vaginosis, especially in patients with frequent recurrence.73 Such manipulation of phage genetic material for effective destruction of bacterial biofilms from biotic or abiotic surfaces may become a feasible option of the 21st century.
Phage-Derived Enzymes
Some enzymes encoded with phages maybe useful for treating bacterial infections and biofilms. Under current safety standards and regulations, the application of phage products is easier than use of the phage itself. In relation to this, two main types of phage degradation enzymes are useful in the removal of biofilms: lysins and depolymerases.
Application of Lysins
Lysins are peptidoglycan hydrolases encoded by Gram-positive infecting phages and include muramidase, transglycosylase, glucosaminidase, amidase, and endopeptidase. The application of endolysins against Gram-negative pathogens is impaired by the presence of a protecting outer membrane layer, however, the combination with membrane permeabilizers turned out to significantly improve lysin efficiency.74,75 More recently, genetic engineering has allowed the design of lysin/cationic peptide combination called Artilysins; the lysin/bacteriocin version to obtain Lysocins, and the lysin/phage receptor binding proteins to generate Innolysins, as an anticipated promising strategy.2
In terms of antibiofilm activity, phage lytic proteins offer interesting properties. In a recent study on investigating the combination of a phage-derived lytic protein, CHAPSH3b, and the virulent bacteriophage phiIPLA-RODI, the results showed that synergy exist between both antimicrobials for the removal of S. aureus biofilms, with greater reductions in viable cell counts observed when phage and lysin were applied together compared to the individual treatments. Time-kill curves and confocal microscopy revealed that the fast antibacterial action of CHAPSH3b reduces the population up to 7 hours after initial exposure, which is subsequently followed by phage predation, limiting regrowth of the bacterial population.76 The amidase domain of phage vB_LmoS_293 was shown to inhibit biofilm formation of L. monocytogenes on abiotic surfaces. S. pyogenes biofilm matrix refractory to conventional antibiotics were readily destroyed by endolysin PlyC with minimum biofilm eradication concentration values of two and four orders of magnitude lower by mass and molarity respectively, than the conventional antibiotics.77 Potent antibiofilm agent, lysin CF-301, removed S. aureus and mixed-species biofilms formed on polystyrene, glass, surgical mesh, and catheters, with an improvement in antibiofilm activity when combined with cell wall hydrolase lysostaphin.78 Similarly, phage endolysin LysCSA13 when applied, showed high efficacy in removing staphylococcal biofilms on various surfaces, including polystyrene, glass, and stainless steel, with about 80–90% decrease in biofilm mass.79 In a study involving Gram-negative bacteria, the endolysin of A. baumannii bacteriophage D2, Abtn-4, was found to have broad antimicrobial activity against MDR S. aureus, P. aeruginosa, K. pneumoniae, Enterococcus and Salmonella in the absence of outer membrane permeabilizers. Abtn-4 had the ability to reduce biofilm formation and showed antimicrobial activity against phage-resistant bacterial mutants.80 With their activities independent of the bacterial physiological state, lysins are relevant for biofilm removal especially phage-resistant bacteria.
Application of Depolymerases
Phage depolymerases are proteins encoded in the region of structural genes in a phage genome which recognize, bind, and digest the polysaccharide compounds of bacterial cell walls. EPSs are mainly responsible for the structural and functional integrity of bacterial biofilms and have an influence on their virulence. Interestingly, Gutiérrez et al81 applied the EPS depolymerase Dpo7, derived from bacteriophage vB_SepiS-phiIPLA7, against staphylococcal biofilms. The study revealed that over 90% of biofilm-attached cells were removed by Dpo7 in all polysaccharide producer staphylococcal strains except polysaccharide-independent biofilm formed by S. aureus V329. Additionally, the pre-treatment of polystyrene surfaces with Dpo7 resulted in the reduction of biofilm biomass by 53–85% in two-thirds of the tested strains. EPS depolymerase Dpo7 has the ability to inhibit biofilm formation and can also disperse biofilms generated by different strains of S. epidermidis and S. aureus.81
Recently, the enzymatic activity of a capsular polysaccharide depolymerase TSP of phage ɸAB6, to degrade A. baumannii biofilm, showed significant inhibition of biofilm formation and degradation of formed biofilms. Additionally, TSP inhibited the colonization of A. baumannii on the surface of Foley catheter sections, indicating that it can be used to prevent the adhesion of A. baumannii biofilms to medical device surfaces.82 In another study, recombinant Dep42, a putative tail fiber protein with depolymerase activity from bacteriophage SH-KP152226, showed specific enzymatic activities in the depolymerization of the K47 capsule of K. pneumoniae and significantly inhibited biofilm formation and/or degrade formed biofilms. The study also showed that Dep42 could enhance polymyxin activity against K. pneumoniae biofilms when used in combination with antibiotics, suggesting that combination of identified novel depolymerases encoded by the phages with antibiotics may represent a promising strategy to combat infections caused by MDR and biofilm-forming bacteria.14
Apart from their individual activities as antibiofilm agents, effective removal of biofilms can be achieved with a combination of lysin and depolymerase. In a study on investigating the efficacy of endolysin LysK and poly-N-acetylglucosamine depolymerase DA7 against staphylococcal biofilms, in addition to LysK and DA7 removing static and dynamic biofilms from polystyrene and glass surfaces at low micromolar and nanomolar concentrations respectively, a combination of the enzymes significantly reduced viable cell counts compared to individual enzyme treatment.83 Besides their ease of application, phage enzymes can offer to serve as potential new candidates of antibiofilm agents and antimicrobial drugs (enzybiotics) more than live phages particularly in the advent of phage-resistant bacteria.
Application of Phages in Combination with Other Alternative Antibiofilms
Sometimes, phage therapy as sole antibiofilm agents to destroy bacterial biofilms is inadequate. So far, other than phage therapy, several novel approaches against bacterial biofilms have been proposed.84 These include surface modification or coating methods,85 the use of anti-matrix agents such as enzymes (DNase I or dispersin B)3,86 and chelators of divalent cations,87 the use of naturally occurring antimicrobial peptides88,89 or their synthetic derivatives,90 and the use of QS inhibitors such as curcumin,91 quercetin,92 halogenated furanone compounds,93 RNA-III-inhibiting peptide, ginseng extract, garlic extract, and pyrrhocoricin or microcin B17.94 Others include nanotechnology with the use of metal-based nanoparticles (silver, gold, titanium, copper or zinc), green nanoparticles and polymer-based nanoparticles,21,95 physical methods such as scrubbing and flushing, disinfection methods using chemical alkali-based and acid-based agents, ethanol, chlorine dioxide, or hydrogen peroxide,94 use of monoclonal antibodies,96,97 and antimicrobial photodynamic therapy (PDT) using light-activated photosensitizers that generate cytotoxic species such as reactive oxygen species (ROS).98,99 To completely eradicate bacterial biofilms, a simultaneous or sequential combination approach of phage therapy with other alternative antibiofilm agents is recommended. The combination therapy is composed of phages and/or phage-derived enzymes with nanoparticles, chemical compounds, antimicrobial peptides, and disinfectants (Table 1).
 |
Table 1 Examples of Combination of Phages or Phage-Derived Products and Antimicrobials Applications Against Bacterial Biofilm Formation |
The combination therapy is mainly based on the use of phages and/or phage-derived products with antibiotics. The synergistic actions of mono phages, phage cocktails, or phage-derived enzymes with antibiotics have revealed successful results. The development of phage-based therapy combined with antibiotics might be an advantageous weapon in the arms race between human and MDR or phage-resistant bacteria. In a study by Henriksen et al,117 single phage treatment of P. aeruginosa PAO1 early biofilm stage reduced up to 98% of biofilm biovolume. However, repeated phage treatment increased the biovolume and remained unaffected by the phage treatment, until a combination treatment of phages with ciprofloxacin caused a significant reduction in the abundance of cells in the biofilm.117 The combination treatment of phage PEV20 with ciprofloxacin in another study enhanced biofilm eradication compared with single treatments.118 Likewise, a combination treatment of phage Sb-1 with antibiotics exerted a synergistic effect in eradicating MRSA biofilm119 while similar results have been reported in other studies.120,121
In the application of phages with antibiotics, consideration must be given to the order of therapeutic administration, as well as nature of their interactions, as these factors may affect the outcome of the therapy. However, to achieve optimum synergy, phage treatment must precede antibiotics, as demonstrated in these studies.122,123 Papaianni and his team107 applied a combinatorial antibiofilm strategy based on the use of fatty acids and the bacteriophage Xccɸ1-hydroxyapatite complex optimized against Xanthomonas campestris mature biofilm. The synergic action of these elements was demonstrated and the efficient removal of X. campestris mature biofilm was also proven in a flow cell system, making the proposed approach an effective solution to enhance plant survival in X. campestris infections.107 Manoharadas and his associates111 demonstrated a concerted action of green synthesized silver nanoparticles and bacteriophages in removing pre-formed S. aureus biofilms from an inert glass surface in a time dependent manner. The results demonstrated, for the first time, the rapid cooperative dispersion of the bacterial biofilm. In addition, the synergistic activity of the nanoparticles and bacteriophages cause the loss of viability of the biofilm entrapped bacterial cells thus preventing establishment of a new infection and subsequent colonization. This work further opens up a platform for the combinational therapeutic approach with a variety of nanoparticles and bacteriophages against mono or poly bacterial biofilm in environmental, industrial or clinical settings.111 Using these combinational therapeutic strategies depends on the intention of preventing or treating biofilms related to device-associated nosocomial infections and biofilms formed on native host tissues. Unfortunately, not all of the alternative strategies can be used to treat infections associated with biofilms formed in a patient’s body, for example, surface coating metals, disinfectants, chemical compounds, and mixtures.
Application of Phages as Vehicles to Deliver Antibiofilm Agents
Phages, in addition to their lytic potential and antibiofilm activities, can be modified to be used as delivery vehicles of cargos such as nucleic acids, nanomaterials, therapeutic drugs and diagnostic probes.67,124 In such cases, temperate phages may be of interest for delivering programmable DNA nucleases associated with CRISPR to reverse antibiotic resistance and destroy plasmids that confer antibiotic resistance.125 In this current view, phages can also be modified to deliver antibiofilm agents for biofilm destruction. For example, an injectable hydrogel capable of encapsulating P. aeruginosa phage and delivering active phage to the site of bone infections was engineered. While retaining the bacteriolytic activity after encapsulation and release from the hydrogel, phage-encapsulating hydrogels, apart from effectively killing host bacteria in both planktonic and biofilm phenotypes in vitro, achieved a 4.7-fold reduction in live P. aeruginosa counts at infection site of murine radial segment infected with P. aeruginosa, compared to phage-free hydrogels at 7 days post implantation.126 In another study, a rare bacteriophage was used as a green route to synthesize gold nanoparticles (AuNPs). In addition to showing antibacterial activity against different bacterial pathogens, phage inspired AuNPs inhibition of about 80% of P. aeruginosa biofilms was reported.127 In using magnetic phage-nanocomposite conjugates (PNCs) to target bacteria in biofilm inner layers for bottom-up eradication, phages PEB1 or PEB2 were covalently conjugated with magnetic colloidal nanoparticle clusters of different sizes. Both small and large PNCs dispersed phages evenly throughout the biofilm bottom, significantly disrupting the biofilm bottom layer and detaching the biofilm within 6 hours, with efficient biofilm removal for dual and multi-species biofilm.128 Phages with broad host range are also conjugated with magnetic colloidal nanoparticle clusters to facilitate biofilm penetration and subsequent removal in established biofilms.129 With the continuous evolution of bacteria, such manipulation of phages with nanoparticles, may more than ever, be needed now in the fight against biofilm forming and/or their associated human infections due to antimicrobial resistant bacteria.
Conclusions
Naturally, most bacteria live in the form of biofilms. Biofilms occurrence on medical device surfaces and human tissues, as well as their tolerance to antibiotics, disinfectants and host immunity, brought them to burden in human lives. As a niche not well covered by currently available antibiotics alone, bacterial biofilms necessitate the search for alternative approaches to completely destruct them. Several novel approaches against bacterial biofilms such as nanotechnology, irrigation techniques, surface modification methods and use of natural peptides have been proposed. Nonetheless, phage-based treatments which include the application of mono lytic phages, phage cocktails, engineered phages, and phage-derived enzymes, appear to be effective in destroying bacterial biofilms. Higher efficacy was, however, perceived with a combination therapy of phages or phage-proteins and other alternative antibiofilm agents, as well as phages conjugated with nanoparticles, as delivery vehicles of these antibiofilm agents. To completely eliminate bacterial biofilms, it is recommended that;
- more knowledge on mechanisms of phage application is still needed to increase world acceptance of phage-based therapies.
- future research should aim to broaden the scope of application of phages by promoting engineered phages as well as their enzymes with other alternative agents such as, nanoparticles and antimicrobial peptides,
- future research should also aim to broaden the scope of application of phages by promoting modified phages as delivery vehicles of other antibiofilm agents, and
- efficacy and safety protocols should be prepared to develop an established phage-based method to eliminate biofilms.
Acknowledgment
The authors express their special thanks of gratitude to Mr. Edem Aglago for his genuinely creative contribution in the design of the schematic diagrams. The authors would like also to thank Ms. Patience Aglago for her technical assistance in the design of the diagrams.
Disclosure
The authors declare no potential conflicts of interest in this work.
References
1. Flemming H, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563–575. doi:10.1038/nrmicro.2016.94
2. Azeredo J, García P, Drulis-Kawa Z. Targeting biofilms using phages and their enzymes. Curr Opin Biotechnol. 2021;68:251–261. doi:10.1016/j.copbio.2021.02.002
3. Łusiak-szelachowska M, Weber-Dąbrowska B, Górski A. Bacteriophages and lysins in biofilm control. Virol Sin. 2020;35(2):125–133. doi:10.1007/s12250-019-00192-3
4. Landry KS, Morey JM, Bharat B, Haney NM, Panesar SS. Biofilms—Impacts on human health and its relevance to space travel. Microorganisms. 2020;8(7):998. doi:10.3390/microorganisms8070998
5. Geredew Kifelew L, Mitchell JG, Speck P. Mini-review: efficacy of lytic bacteriophages on multispecies biofilms. Biofouling. 2019;35(4):472–481. doi:10.1080/08927014.2019.1613525
6. Guo Z, Lin H, Ji X, et al. Therapeutic applications of lytic phages in human medicine. Microb Pathog. 2020;142:104048. doi:10.1016/j.micpath.2020.104048
7. Melo LDR, Azevedo NF. New insights on biofilm antimicrobial strategies. Antibiotics. 2021;10(4):407. doi:10.3390/antibiotics10040407
8. Doub JB. Bacteriophage therapy for clinical biofilm infections: parameters that influence treatment protocols and current treatment approaches. Antibiotics. 2020;9(11):799. doi:10.3390/antibiotics9110799
9. Górski A, Borysowski J, Międzybrodzki R. Phage therapy: towards a Successful Clinical Trial. Antibiotics. 2020;9(11):827. doi:10.3390/antibiotics9110827
10. Taha OA, Connerton PL, Connerton IF, El-Shibiny A. Bacteriophage ZCKP1: a potential treatment for Klebsiella pneumoniae isolated from diabetic foot patients. Front Microbiol. 2018;9:2127. doi:10.3389/fmicb.2018.02127
11. Oliveira VC, Macedo AP, Melo LDR, et al. Bacteriophage cocktail-mediated inhibition of Pseudomonas aeruginosa biofilm on endotracheal tube surface. Antibiotics. 2021;10(1):78. doi:10.3390/antibiotics10010078
12. Yang D, Chen Y, Sun E, Hua L, Peng Z, Wu B. Characterization of a lytic bacteriophage vB_EfaS_PHB08 harboring endolysin Lys08 against Enterococcus faecalis biofilms. Microorganisms. 2020;8(9):1332. doi:10.3390/microorganisms8091332
13. Gudina I, Gizachew Z, Woyessa D, Tefera TK. Isolation of bacteriophage and assessment of its activity against biofilms of uropathogenic Escherichia coli in Jimma town, south western Ethiopia. Am J Curr Microbiol. 2018;6(1):52–66.
14. Wu Y, Wang R, Xu M, et al. A Novel polysaccharide depolymerase encoded by the Phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front Microbiol. 2019;10:2768. doi:10.3389/fmicb.2019.02768
15. Maszewska A, Zygmunt M, Grzejdziak I, Różalski A. Use of polyvalent bacteriophages to combat biofilm of Proteus mirabilis causing catheter-associated urinary tract infections. J Appl Microbiol. 2018;125(5):1253–1265. doi:10.1111/jam.14026
16. Srinivasan R, Santhakumari S, Poonguzhali P, Geetha M, Dyavaiah M, Xiangmin L. Bacterial biofilm inhibition: a focused review on recent therapeutic strategies for combating the biofilm mediated infections. Front Microbiol. 2021;12:676458. doi:10.3389/fmicb.2021.676458
17. Herce-Ros N, Álvarez-sagües A, Álvarez-losa L, et al. Antibacterial ability of sodium hypochlorite activated with PUI vs. XPF File against Bacteria growth on Enterococcus faecalis mature biofilm. Dentistry J. 2021;9(6):67. doi:10.3390/dj9060067
18. Tanaka T, Yahata Y, Handa K, et al. An experimental intraradicular biofilm model in the pig for evaluating irrigation techniques. BMC Oral Health. 2021;21(1):177. doi:10.1186/s12903-021-01536-w
19. Topka-Bielecka G, Dydecka A, Necel A, et al. Bacteriophage-derived depolymerases against bacterial biofilm. Antibiotics. 2021;10(2):175. doi:10.3390/antibiotics10020175
20. Jakubovics NS, Goodman SD, Mashburn‐Warren L, Stafford GP, Cieplik F. The dental plaque biofilm matrix. Darveau RP, Curtis MAeds. Periodontology. 2021;86(1):32–56. doi:10.1111/prd.12361
21. Fulaz S, Vitale S, Quinn L, Casey E. Nanoparticle–biofilm interactions: the role of the EPS matrix. Trends Microbiol. 2019;27(11):915–926. doi:10.1016/j.tim.2019.07.004
22. Dragoš A, Kovács ÁT. The peculiar functions of the bacterial extracellular matrix. Trends Microbiol. 2017;25(4):257–266. doi:10.1016/j.tim.2016.12.010
23. Guła G, Dorotkiewicz-Jach A, Korzekwa K, Valvano MA, Drulis-Kawa Z. Complex signaling networks controlling dynamic molecular changes in Pseudomonas aeruginosa biofilm. Curr Med Chem. 2019;26(11):1979–1993. doi:10.2174/0929867325666180912110151
24. Abebe GM. The Role of bacterial biofilm in antibiotic resistance and food contamination. Int J Microbiol. 2020;2020:170814. doi:10.1155/2020/1705814
25. Brindhadevi K, LewisOscar F, Mylonakis E, Shanmugam S, Verma TN, Pugazhendhi A. Biofilm and quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochemistry. 2020;96:49–57. doi:10.1016/j.procbio.2020.06.001
26. Prescott RD, Decho AW. Flexibility and adaptability of quorum sensing in nature. Trends Microbiol. 2020;28(6):436–444. doi:10.1016/j.tim.2019.12.004
27. Tian F, Li J, Nazir A, Tong Y. Bacteriophage – a promising alternative measure for bacterial biofilm control. Infect Drug Resist. 2021;14:205–217. doi:10.2147/IDR.S290093
28. Jamal M, Ahmad W, Andleeb S, et al. Bacterial biofilm and associated infections. J Chin Med Assoc. 2018;81(1):7–11. doi:10.1016/j.jcma.2017.07.012
29. Karygianni L, Ren Z, Koo H, Thurnheer T. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 2020;28(8):668–681. doi:10.1016/j.tim.2020.03.016
30. Toyofuku M, Inaba T, Kiyokawa T, Obana N, Yawata Y, Nomura N. Environmental factors that shape biofilm formation. Biosci Biotechnol Biochem. 2016;80(1):7–12. doi:10.1080/09168451.2015.1058701
31. Moormeier DE, Bayles KW. Staphylococcus aureus biofilm: a complex developmental organism. Mol Microbiol. 2017;104(3):365–376. doi:10.1111/mmi.13634
32. Rumbaugh KP, Sauer K. Biofilm dispersion. Nat Rev Microbiol. 2020;18(10):571–586. doi:10.1038/s41579-020-0385-0
33. Flemming HC, Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 2019;17(4):247–260. doi:10.1038/s41579-019-0158-9
34. Reza A, Sutton JM, Rahman KM. Effectiveness of efflux pump inhibitors as biofilm disruptors and resistance breakers in Gram-negative (ESKAPEE) bacteria. Antibiotics. 2019;8(4):229. doi:10.3390/antibiotics8040229
35. Percival SL, Suleman L, Vuotto C, Donelli G. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J Med Microbiol. 2015;64(4):323–334. doi:10.1099/jmm.0.000032
36. Li X-H, Lee J-H. Antibiofilm agents: a new perspective for antimicrobial strategy. J Microbiol. 2017;55(10):753–766. doi:10.1007/s12275-017-7274-x
37. Batinovic S, Wassef F, Knowler SA, et al. Bacteriophages in natural and artificial environments. Pathogens. 2019;8(3):100. doi:10.3390/pathogens8030100
38. Kassa T. Bacteriophages against pathogenic bacteria and possibilities for future application in Africa. Infect Drug Resist. 2021;14:17–31. doi:10.2147/IDR.S284331
39. Gutiérrez D, Rodríguez-Rubio L, Martínez B, Rodríguez A, García P. Bacteriophages as weapons against bacterial biofilms in the food Industry. Front Microbiol. 2016;7:825. doi:10.3389/fmicb.2016.00825
40. González S, Fernández L, Gutiérrez D, Campelo AB, Rodríguez A, García P. Analysis of different parameters affecting diffusion, propagation and survival of Staphylophages in bacterial biofilms. Front Microbiol. 2018;9:2348. doi:10.3389/fmicb.2018.02348
41. Yang D, Wang Z, Ma J, et al. Glycine cleavage system and cAMP receptor protein co-regulate CRISPR/cas3 expression to resist bacteriophage. Viruses. 2020;12(1):90. doi:10.3390/v12010090
42. Gutiérrez D, Fernández L, Martínez B, Ruas-Madiedo P, García P, Rodríguez A. Real-Time assessment of Staphylococcus aureus biofilm disruption by phage-derived proteins. Front Microbiol. 2017;8:1632. doi:10.3389/fmicb.2017.01632
43. Pires DP, Melo LDR, Vilas Boas D, Sillankorva S, Azeredo J. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr Opin Microbiol. 2017;39:48–56. doi:10.1016/j.mib.2017.09.004
44. Guerrero-Ferreira RC, Viollier PH, Ely B, et al. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proce National Acad Sci. 2011;108(24):9963–9968. doi:10.1073/pnas.1012388108
45. Azam AH, Tanji Y. Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl Microbiol Biotechnol. 2019;103(5):2121–2131. doi:10.1007/s00253-019-09629-x
46. Davies EV, James CE, Williams D, et al. Temperate phages both mediate and drive adaptive evolution in pathogen biofilms. Proce National Acad Sci. 2016;113(29):8266–8271. doi:10.1073/pnas.1520056113
47. Fernández L, González S, Campelo AB, Martínez B, Rodríguez A, García P. Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus. Sci Rep. 2017;7(1):40965. doi:10.1038/srep40965
48. Secor PR, Sweere JM, Michaels LA, et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe. 2015;18(5):549–559. doi:10.1016/j.chom.2015.10.013
49. Drulis-Kawa Z, Maciejewska B. Special Issue: “Bacteriophages and Biofilms”. Viruses. 2021;13(2):257. doi:10.3390/v13020257
50. Loponte R, Pagnini U, Iovane G, Pisanelli G. Phage therapy in veterinary medicine. Antibiotics. 2021;10(4):421. doi:10.3390/antibiotics10040421
51. Rakov C, Ben Porat S, Alkalay-Oren S, et al. Targeting biofilm of MDR Providencia stuartii by phages using a catheter model. Antibiotics. 2021;10(4):375. doi:10.3390/antibiotics10040375
52. Ben-Zaken H, Kraitman R, Coppenhagen-Glazer S, et al. Isolation and characterization of Streptococcus mutans phage as a possible treatment agent for caries. Viruses. 2021;13(5):825. doi:10.3390/v13050825
53. Rajabi Z, Kermanshahi R, Dallal SMM, Erfani Y, Ranjbar R. Isolation of the bacteriophages inhibiting the expression of the genes involved in biofilm formation by Streptococcus mutans. Jundishapur J Microbiol. 2021;14(1):e113206. doi:10.5812/jjm.113206.Research
54. Magin V, Garrec N, Andrés Y. Selection of bacteriophages to control in vitro 24 h old biofilm of Pseudomonas aeruginosa isolated from drinking and thermal water. Viruses. 2019;11(8):749. doi:10.3390/v11080749
55. Kim SG, Giri SS, Yun S, et al. Two novel bacteriophages control multidrug- and methicillin-resistant Staphylococcus pseudintermedius biofilm. Front Med. 2021;8:524059. doi:10.3389/fmed.2021.524059
56. Topka-Bielecka G, Nejman-Faleńczyk B, Bloch S, et al. Phage-bacteria interactions in potential applications of bacteriophage vB_EfaS-271 against Enterococcus faecalis. Viruses. 2021;13(2):318. doi:10.3390/v13020318
57. Song J, Ruan H, Chen L, et al. Potential of bacteriophages as disinfectants to control of Staphylococcus aureus biofilms. BMC Microbiol. 2021;21(1):57. doi:10.1186/s12866-021-02117-1
58. Montso PK, Mlambo V, Ateba CN. Efficacy of novel phages for control of multi-drug resistant Escherichia coli O177 on artificially contaminated beef and their potential to disrupt biofilm formation. Food Microbiol. 2021;94:103647. doi:10.1016/j.fm.2020.103647
59. Nale JY, Clokie MR. Preclinical data and safety assessment of phage therapy in humans. Curr Opin Biotechnol. 2021;68:310–317. doi:10.1016/j.copbio.2021.03.002
60. Zeid AAA, Swelim MA, Reda FM, Abd El Haveez A, Nasr-Eldin M. Effectiveness of four lytic phages against biofilm-producing and multidrug-resistant Escherichia coli. Benha J Applied Sci. 2021;6(1):53–65. doi:10.21608/bjas.2021.167708
61. Zurabov F, Zhilenkov E. Characterization of four virulent Klebsiella pneumoniae bacteriophages, and evaluation of their potential use in complex phage preparation. Virol J. 2021;18(1):9. doi:10.1186/s12985-020-01485-w
62. Askoura M, Saed N, Enan G, Askora A. Characterization of polyvalent bacteriophages targeting multidrug-resistant Klebsiella pneumonia with enhanced anti-biofilm activity. App Biochem Microbiol. 2021;57(1):117–126. doi:10.1134/S000368382101004X
63. Tkhilaishvili T, Winkler T, Müller M, Perka C, Trampuz A. Bacteriophages as adjuvant to antibiotics for the treatment of periprosthetic joint infection caused by multidrug-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2020;64(1):e00924–19. doi:10.1128/AAC.00924-19
64. Abdurahman MA, Tosun I, Durukan I, Khorshidtalab M, Kilic AO. Four Temperate bacteriophages from methicillin-resistant Staphylococcus aureus show broad bactericidal and biofilm removal activities. Kafkas Univ Veteriner Fakultesi Dergisi. 2021;27:1.
65. Kifelew LG, Warner MS, Morales S, et al. Efficacy of lytic phage cocktails on Staphylococcus aureus and Pseudomonas aeruginosa in mixed-species planktonic cultures and biofilms. Viruses. 2020;12(5):559. doi:10.3390/v12050559
66. Gutiérrez D, Vandenheuvel D, Martínez B, Rodríguez A, Lavigne R, García P. Two phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono- and dual-species staphylococcal biofilms. Appl Environ Microbiol. 2015;81(10):3336–3348. doi:10.1128/AEM.03560-14
67. Peng H, Chen IA. Phage engineering and the evolutionary arms race. Curr Opin Biotechnol. 2021;68:23–29. doi:10.1016/j.copbio.2020.09.009
68. Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proce National Acad Sci. 2007;104(27):11197–11202. doi:10.1073/pnas.0704624104
69. Chen J, Chen Z, Yuan K, Huang Z, Mao M. Recombinant bacteriophage T4 Rnl1 impacts Streptococcus mutans biofilm formation. J Oral Microbiol. 2021;13(1):1860398. doi:10.1080/20002297.2020.1860398
70. Pei R, Lamas-Samanamud GR. Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Appl Environ Microbiol. 2014;80(17):5340–5348. doi:10.1128/AEM.01434-14
71. Guo D, Chen J, Zhao X, et al. Genetic and chemical engineering of phages for controlling multidrug-resistant bacteria. Antibiotics. 2021;10(2):202. doi:10.3390/antibiotics10020202
72. Payaslian F, Gradaschi V, Piuri M. Genetic manipulation of phages for therapy using BRED. Curr Opin Biotechnol. 2021;68:8–14. doi:10.1016/j.copbio.2020.09.005
73. Landlinger C, Tisakova L, Oberbauer V, et al. Engineered phage endolysin eliminates Gardnerella biofilm without damaging beneficial bacteria in bacterial vaginosis Ex Vivo. Pathogens. 2021;10(1):54. doi:10.3390/pathogens10010054
74. Huang Z, Zhang Z, Tong J, et al. Phages and their lysins: toolkits in the battle against foodborne pathogens in the postantibiotic era. Comprehensive Rev Food Sci Food Safety. 2021:2:1–25. doi:10.1111/1541-4337.12757
75. Pinto AM, Silva MD, Pastrana LM, Bañobre-López M, Sillankorva S. The clinical path to deliver encapsulated phages and lysins. FEMS Microbiol Rev. 2021;19:1–29. doi:10.1093/femsre/fuab019
76. Duarte AC, Fernández L, De Maesschalck V, et al. Synergistic action of phage phiIPLA-RODI and lytic protein CHAPSH3b: a combination strategy to target Staphylococcus aureus biofilms. Npj Biofilms Microbiomes. 2021;7(1):39. doi:10.1038/s41522-021-00208-5
77. Shen Y, Köller T, Kreikemeyer B, Nelson DC. Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage-encoded endolysin. J Antimicrobial Chemotherapy. 2013;68(8):1818–1824. doi:10.1093/jac/dkt104
78. Schuch R, Khan BK, Raz A, Rotolo JA, Wittekind M. Bacteriophage lysin CF-301, a potent antistaphylococcal biofilm agent. Antimicrob Agents Chemother. 2017;61(7):e02666–16. doi:10.1128/AAC.02666-16
79. Cha Y, Son B, Ryu S. Effective removal of staphylococcal biofilms on various food contact surfaces by Staphylococcus aureus phage endolysin LysCSA13. Food Microbiol. 2019;84:103245. doi:10.1016/j.fm.2019.103245
80. Yuan Y, Li X, Wang L, et al. The endolysin of the Acinetobacter baumannii phage vB_AbaP_D2 shows broad antibacterial activity. Microb Biotechnol. 2021;14(2):403–418. doi:10.1111/1751-7915.13594
81. Gutiérrez D, Briers Y, Rodríguez-Rubio L, et al. Role of the pre-neck appendage protein (Dpo7) from phage vB_SepiS-phiIPLA7 as an anti-biofilm agent in Staphylococcal species. Front Microbiol. 2015;6:1315. doi:10.3389/fmicb.2015.01315
82. Shahed-Al-Mahmud M, Roy R, Sugiokto FG, et al. Phage φAB6-borne depolymerase combats Acinetobacter baumannii biofilm formation and infection. Antibiotics. 2021;10(3):279. doi:10.3390/antibiotics10030279
83. Olsen N, Thiran E, Hasler T, et al. Synergistic removal of static and dynamic Staphylococcus aureus biofilms by combined treatment with a bacteriophage endolysin and a polysaccharide depolymerase. Viruses. 2018;10(8):438. doi:10.3390/v10080438
84. Li X, Sun L, Zhang P, Wang Y. Novel approaches to combat medical device-associated biofilms. Coatings. 2021;11(3):294. doi:10.3390/coatings11030294
85. Wang Y, Jayan G, Patwardhan D, Phillips SK. Antimicrobial and anti-biofilm medical devices: public health and regulatory science challenges. In: Zhang Z, Wagner VE editors. Antimicrobial coatings and modifications on medical devices. Springer International; 2017:1–273. doi:10.1007/978-3-319-57494-3.
86. Waryah CB, Wells K, Ulluwishewa D, et al. In Vitro antimicrobial efficacy of tobramycin against Staphylococcus aureus biofilms in combination with or without DNase I and/or Dispersin B: a preliminary investigation. Microbial Drug Resistance. 2017;23(3):384–390. doi:10.1089/mdr.2016.0100
87. Cavaliere R, Ball JL, Turnbull L, Whitchurch CB. The biofilm matrix destabilizers, EDTA and DNaseI, enhance the susceptibility of nontypeable Hemophilus influenzae biofilms to treatment with ampicillin and ciprofloxacin. MicrobiologyOpen. 2014;3(4):557–567. doi:10.1002/mbo3.187
88. Shahrour H, Ferrer-espada R, Dandache I, et al. AMPs as anti-biofilm agents for human therapy and prophylaxis. Matsuzaki K, editor. Antimicrobial Peptides;Vol. 1117. Springer Singapore; 2019. 257–279. doi:10.1007/978-981-13-3588-4
89. Kazemzadeh-Narbat M, Cheng H, Chabok R, et al. Strategies for antimicrobial peptide coatings on medical devices: a review and regulatory science perspective. Crit Rev Biotechnol. 2020;41(1):94–120. doi:10.1080/07388551.2020.1828810
90. De La Fuente-núñez C, Cardoso MH, De Souza Cândido E, Franco OL, Hancock REW. Synthetic antibiofilm peptides. Biochimica et Biophysica Acta. 2016;1858(5):1061–1069. doi:10.1016/j.bbamem.2015.12.015
91. Zhao X, Zhao F, Zhong N. Quorum sensing inhibition and anti-biofilm activity of traditional Chinese medicines. Food Safety. 2018;1:37–50. doi:10.5772/intechopen.74658
92. Gopu V, Meena CK, Shetty PH. Quercetin influences quorum sensing in food borne bacteria: in-vitro and in-silico evidence. PLoS One. 2015;10(8):e0134684. doi:10.1371/journal.pone.0134684
93. Zhang W, Li C. Exploiting quorum sensing interfering strategies in gram-negative bacteria for the enhancement of environmental applications. Front Microbiol. 2016;6:1535. doi:10.3389/fmicb.2015.01535
94. Roy R, Tiwari M, Donelli G, Tiwari V. Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence. 2018;9(1):522–554. doi:10.1080/21505594.2017.1313372
95. Zagami R, Franco D, Pipkin JD, et al. Sulfobutylether-β-cyclodextrin/5,10,15,20-tetrakis(1-methylpyridinium-4-yl)porphine nanoassemblies with sustained antimicrobial phototherapeutic action. Int J Pharm. 2020;585:119487. doi:10.1016/j.ijpharm.2020.119487
96. Tursi SA, Puligedda RD, Szabo P, et al. Salmonella Typhimurium biofilm disruption by a human antibody that binds a pan-amyloid epitope on curli. Nat Commun. 2020;11(1):1007. doi:10.1038/s41467-020-14685-3
97. Raafat D, Otto M, Reppschläger K, Iqbal J, Holtfreter S. Fighting Staphylococcus aureus biofilms with monoclonal antibodies. Trends Microbiol. 2019;27(4):303–322. doi:10.1016/j.tim.2018.12.009
98. Yuan Z, Tao B, He Y, et al. Remote eradication of biofilm on titanium implant via near-infrared light triggered photothermal/photodynamic therapy strategy. Biomaterials. 2019;223:119479. doi:10.1016/j.biomaterials.2019.119479
99. Tan L, Li J, Liu X, et al. Rapid biofilm eradication on bone implants using red phosphorus and near-infrared light. Adv Mater. 2018;30(31):1801808. doi:10.1002/adma.201801808
100. Grygorcewicz B, Wojciuk B, Roszak M, et al. Environmental phage-based cocktail and antibiotic combination effects on Acinetobacter baumannii biofilm in a human urine model. Microbial Drug Resistance. 2021;27(1):25–35. doi:10.1089/mdr.2020.0083
101. Maszewska A, Moryl M, Wu J, Liu B, Feng L, Rozalski A. Amikacin and bacteriophage treatment modulates outer membrane proteins composition in Proteus mirabilis biofilm. Sci Rep. 2021;11(1):1522. doi:10.1038/s41598-020-80907-9
102. Tkhilaishvili T, Wang L, Perka C, Trampuz A, Gonzalez Moreno M. Using bacteriophages as a Trojan Horse to the killing of dual-species biofilm formed by Pseudomonas aeruginosa and methicillin resistant Staphylococcus aureus. Front Microbiol. 2020;11:695. doi:10.3389/fmicb.2020.00695
103. Shlezinger M, Coppenhagen-Glazer S, Gelman D, Beyth N, Hazan R. Eradication of vancomycin-resistant Enterococci by combining phage and vancomycin. Viruses. 2019;11(10):954. doi:10.3390/v11100954
104. Sagar SS, Kumar R, Kaistha SD. Efficacy of phage and ciprofloxacin co-therapy on the formation and eradication of Pseudomonas aeruginosa biofilms. Arab J Sci Eng. 2017;42(1):95–103. doi:10.1007/s13369-016-2194-3
105. Davis CM, McCutcheon JG, Dennis JJ. Aztreonam lysine increases the activity of phages E79 and phiKZ against Pseudomonas aeruginosa PA01. Microorganisms. 2021;9(1):152. doi:10.3390/microorganisms9010152
106. Lu Y, Wang Y, Wang J, et al. Phage Endolysin LysP108 Showed Promising Antibacterial Potential Against Methicillin-resistant Staphylococcus aureus. Front Cell Infect Microbiol. 2021;11:668430. doi:10.3389/fcimb.2021.668430
107. Papaianni M, Ricciardelli A, Casillo A, et al. The union is strength: the synergic action of long fatty acids and a bacteriophage against Xanthomonas campestris biofilm. Microorganisms. 2020;9(1):60. doi:10.3390/microorganisms9010060
108. Chhibber S, Nag D, Bansal S. Inhibiting biofilm formation by Klebsiella pneumoniae B5055 using an iron antagonizing molecule and a bacteriophage. BMC Microbiol. 2013;13(1):174. doi:10.1186/1471-2180-13-174
109. Zhang Y, Hu Z. Combined treatment of Pseudomonas aeruginosa biofilms with bacteriophages and chlorine. Biotechnol Bioeng. 2013;110(1):286–295. doi:10.1002/bit.24630
110. Oliveira A, Sousa JC, Silva AC, Melo LDR, Sillankorva S. Chestnut honey and bacteriophage application to control Pseudomonas aeruginosa and Escherichia coli biofilms: evaluation in an ex vivo wound model. Front Microbiol. 2018;9:1725. doi:10.3389/fmicb.2018.01725
111. Manoharadas S, Altaf M, Alrefaei AF, Devasia RM, Badjah Hadj AYM, Abuhasil MSA. Concerted dispersion of Staphylococcus aureus biofilm by bacteriophage and ‘green synthesized’ silver nanoparticles. RSC Adv. 2021;11(3):1420–1429. doi:10.1039/D0RA09725J
112. Chai Z, Wang J, Tao S, Mou H. Application of bacteriophage-borne enzyme combined with chlorine dioxide on controlling bacterial biofilm. Food Sci Technol. 2014;59(2):1159–1165. doi:10.1016/j.lwt.2014.06.033
113. Lemon DJ, Kay MK, Titus JK, et al. Construction of a genetically modified T7Select phage system to express the antimicrobial peptide 1018. J Microbiol. 2019;57(6):532–538. doi:10.1007/s12275-019-8686-6
114. Yüksel FN, Buzrul S, Akçelik M, Akçelik N. Inhibition and eradication of Salmonella Typhimurium biofilm using P22 bacteriophage, EDTA and nisin. Biofouling. 2018;34(9):1046–1054. doi:10.1080/08927014.2018.1538412
115. Duc HM, Son HM, Ngan PH, et al. Isolation and application of bacteriophages alone or in combination with nisin against planktonic and biofilm cells of Staphylococcus aureus. Appl Microbiol Biotechnol. 2020;104(11):5145–5158. doi:10.1007/s00253-020-10581-4
116. Ni P, Wang L, Deng B, et al. Combined application of bacteriophages and carvacrol in the control of Pseudomonas syringae pv. actinidiae planktonic and biofilm forms. Microorganisms. 2020;8(6):837. doi:10.3390/microorganisms8060837
117. Henriksen K, Rørbo N, Rybtke ML, et al. P. aeruginosa flow-cell biofilms are enhanced by repeated phage treatments but can be eradicated by phage–ciprofloxacin combination:-monitoring the phage-P. aeruginosa biofilms interactions. Pathog Dis. 2019;77(2):ftz011. doi:10.1093/femspd/ftz011
118. Chang RYK, Das T, Manos J, Kutter E, Morales S, Chan H-K. Bacteriophage PEV20 and ciprofloxacin combination treatment enhances removal of Pseudomonas aeruginosa biofilm isolated from cystic fibrosis and wound patients. AAPS J. 2019;21(3):49. doi:10.1208/s12248-019-0315-0
119. Tkhilaishvili T, Lombardi L, Klatt A-B, Trampuz A, Di Luca M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int J Antimicrob Agents. 2018;52(6):842–853. doi:10.1016/j.ijantimicag.2018.09.006
120. Kolenda C, Josse J, Medina M, et al. Evaluation of the activity of a combination of three bacteriophages alone or in association with antibiotics on Staphylococcus aureus embedded in biofilm or internalized in osteoblasts. Antimicrob Agents Chemother. 2020;64(3):e02231–19. doi:10.1128/AAC.02231-19
121. Nouraldin AAM, Baddour MM, Harfoush RAH, Essa SAM. Bacteriophage-antibiotic synergism to control planktonic and biofilm producing clinical isolates of Pseudomonas aeruginosa. Alexandria J Med. 2016;52(2):99–105. doi:10.1016/j.ajme.2015.05.002
122. Kumaran D, Taha M, Yi Q, et al. Does treatment order matter? Investigating the ability of bacteriophage to augment antibiotic activity against Staphylococcus aureus biofilms. Front Microbiol. 2018;9:127. doi:10.3389/fmicb.2018.00127
123. Chaudhry WN, Concepción-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. Rozen DEed. PLoS One. 2017;12(1):e0168615. doi:10.1371/journal.pone.0168615
124. Łobocka M, Dąbrowska K, Górski A. Engineered bacteriophage therapeutics: rationale, challenges and future. BioDrugs. 2021;35(3):255–280. doi:10.1007/s40259-021-00480-z
125. Lenneman BR, Fernbach J, Loessner MJ, Lu TK, Kilcher S. Enhancing phage therapy through synthetic biology and genome engineering. Curr Opin Biotechnol. 2021;68:151–159. doi:10.1016/j.copbio.2020.11.003
126. Wroe JA, Johnson CT, García AJ. Bacteriophage delivering hydrogels reduce biofilm formation in vitro and infection in vivo. J Biomed Mater Res A. 2020;108(1):39–49. doi:10.1002/jbm.a.36790
127. Ahiwale SS, Bankar AV, Tagunde S, Kapadnis BP. Bacteriophage mediatedgold nanoparticles synthesis and their anti-biofilm activity. Indian J Microbiol. 2017;57(2):188–194. doi:10.1007/s12088-017-0640-x
128. Yu P, Wang Z, Marcos-Hernandez M, et al. Bottom-up biofilm eradication using bacteriophage-loaded magnetic nanocomposites: a computational and experimental study. Environ Sci. 2019;6(12):3539–3550. doi:10.1039/C9EN00827F
129. Li LL, Yu P, Wang X, et al. Enhanced biofilm penetration for microbial control by polyvalent phages conjugated with magnetic colloidal nanoparticle clusters (CNCs). Environ Sci. 2017;4(9):1817–1826. doi:10.1039/c7en00414a
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.