Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 8
Asthma pharmacogenetics and the development of genetic profiles for personalized medicine
Authors Ortega V, Meyers D, Bleecker E
Received 29 June 2014
Accepted for publication 30 August 2014
Published 16 January 2015 Volume 2015:8 Pages 9—22
DOI https://doi.org/10.2147/PGPM.S52846
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin Bluth
Victor E Ortega, Deborah A Meyers, Eugene R Bleecker
Center for Genomics and Personalized Medicine Research, Pulmonary Medicine, Wake Forest School of Medicine, Winston-Salem, NC, USA
Abstract: Human genetics research will be critical to the development of genetic profiles for personalized or precision medicine in asthma. Genetic profiles will consist of gene variants that predict individual disease susceptibility and risk for progression, predict which pharmacologic therapies will result in a maximal therapeutic benefit, and predict whether a therapy will result in an adverse response and should be avoided in a given individual. Pharmacogenetic studies of the glucocorticoid, leukotriene, and β2-adrenergic receptor pathways have focused on candidate genes within these pathways and, in addition to a small number of genome-wide association studies, have identified genetic loci associated with therapeutic responsiveness. This review summarizes these pharmacogenetic discoveries and the future of genetic profiles for personalized medicine in asthma. The benefit of a personalized, tailored approach to health care delivery is needed in the development of expensive biologic drugs directed at a specific biologic pathway. Prior pharmacogenetic discoveries, in combination with additional variants identified in future studies, will form the basis for future genetic profiles for personalized tailored approaches to maximize therapeutic benefit for an individual asthmatic while minimizing the risk for adverse events.
Keywords: asthma, pharmacogenetics, response heterogeneity, single nucleotide polymorphism, genome-wide association study
Introduction
Asthma is a common, inflammatory airways disease that impacts more than 300 million people worldwide. There are multiple therapies available, which target different biologic pathways for the management of asthma, including leukotriene modifiers, glucocorticoids, inhaled corticosteroids (ICS), β2-adrenergic receptor agonists and anticholinergics, theophylline, cromones, and monoclonal antibodies to immunoglobulin E. Despite treatment with a combination of these therapies, a subgroup of refractory asthmatics will experience poor symptom control and exacerbations.1,2 In addition, specific therapies have been associated with adverse and sometimes life-threatening responses. These therapies target biologic pathways consisting of multiple candidate genes with variation that may alter the therapeutic response, thus providing the basis for development of genetic biomarker panels or genetic profiles for personalized or precision medicine.3–5
Genetic studies of asthma susceptibility, severity, and responsiveness to different pharmacologic therapies have been published, are ongoing, and will lead the way to development of genetic profiles. The ideal genetic profiles will consist of gene variants that identify individual disease susceptibility and risk for progression to more severe disease, predict which pharmacologic therapies will result in a maximal therapeutic benefit, or predict whether a therapy will result in an adverse response and should be avoided in a given individual.6 Genetic studies evaluating the effect of gene variants on beneficial or adverse responses to pharmacologic therapies are referred to as pharmacogenetic studies.
Pharmacogenetics and genetic profiles for personalized medicine
Pharmacogenetics is an important focus of asthma genetics research that evaluates a gene-by-environment interaction where environment is the exposure to a pharmacologic therapy and the outcome of interest is phenotype alteration in response to the therapy (which may include adverse effect phenotypes, Figure 1). The heritability of pharmacologic responses was initially observed in twin-based studies and clinical trials showing that intraindividual (within the same individual) responses to particular therapies vary much less than interindividual (between different individuals) responsiveness.7,8 Pharmacogenetic studies of the glucocorticoid, leukotriene, and β2-adrenergic receptor pathways have primarily focused on candidate genes within these pathways and have identified genetic loci associated with therapeutic responsiveness in asthma (Tables 1–3).
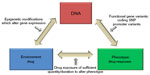  | Figure 1 Premise of pharmacogenetics research in complex diseases. |
Most asthma pharmacogenetic studies evaluate pharmacodynamic endpoints such as lung function, symptom severity, and asthma exacerbation frequency. These predetermined trial endpoints are usually analyzed for genetic associations after completion of a clinical trial; however, a minority of studies use a prospective, genotype-stratified approach where DNA is collected and genotyped for a variant of interest before trial recruitment and forms the basis for randomization to drug or placebo.9 A prospective genotype-stratified approach had the advantage of being able to ensure sufficient statistical power to analyze less common variants in a population since recruitment is based on a risk gene variant. In contrast, large clinical trial cohorts with DNA collection have allowed for genome-wide association studies (GWAS) and other novel methodologies that have taken advantage of high-throughput genome-wide scanning methods to identify novel pharmacogenetic loci.
Pharmacogenetic loci identified through biologic candidate gene studies and GWAS likely interact with each other and interact with other as yet unidentified gene variants to influence therapeutic responsiveness to different pharmacologic agents and the risk for adverse responses. The interaction between different gene variants in determining phenotypic variability in asthma is consistent with multi-gene models showing that an increased number of lung function gene variants has been associated with an increased frequency of severe asthma and lung function abnormalities.10,11 These gene variants in combination with additional variants yet to be identified will form the basis of genetic profiles for personalized or precision medicine in asthma, a complex disease with multiple genetic and environmental factors. Predictive genetic profiles for personalized therapeutic interventions are becoming a reality in the management of lung cancer (an acquired somatic genetic disease) and cystic fibrosis (an autosomal recessive genetic disease), but are not yet applicable to asthma management.12,13
In this review, we summarize prior pharmacogenetic discoveries of common genetic variants identified in candidate gene studies and in a small number of GWAS. We also discuss the issue of lost heritability or missing genetic factors not yet accounted for by prior studies. This is being addressed through targeted analyses of gene–gene interactions, the role of rare variants, ancestry-based genetic studies, and identification of novel candidate genes through gene expression studies. While much of our discussion is aimed at biologic pathways targeted by commonly used asthma therapies, we also summarize how pharmacogenetic research has identified genes that could impact response to biologic therapies currently under development that target specific biologic pathways.
Glucocorticoid pathway
Glucocorticoids are the most commonly used and most effective first-line therapy for the management of asthma and can be administered as an oral or injectable systemic therapy during an exacerbation or as a chronic controller therapy with ICS.14,15 The majority of asthmatics show improvements in lung function in response to ICS therapy; however, there is a small subset that shows minimal or negative lung function responsiveness.16 These individuals with lack of responsiveness to ICS are classified as steroid-resistant or refractory, and continue to be symptomatic or experience asthma exacerbations on optimal ICS therapy (after noncompliance is eliminated).1,2
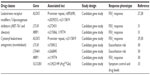
Glucocorticoids target a biologic pathway consisting of biosynthetic hormones that bind and activate a cytosolic chaperone-receptor heterocomplex in the cytosol which translocates into the nucleus to repress the transcription of proinflammatory genes and enhance the transcription of anti-inflammatory genes. Pharmacogenetic studies of the glucocorticoid pathway were initially based on biologic candidate genes encoding the glucocorticoid biosynthetic pathway, the receptor heterocomplex, and chaperone proteins (Table 1). A biologic candidate gene study of the corticotropin-releasing hormone gene (CRHR1) in 1,117 asthmatics randomized to ICS therapy from three clinical trial cohorts identified two CRHR1 single nucleotide polymorphisms (SNPs, denoted by a reference sequence [rs] number or the coding change when applicable) associated with lung function response.17 A subsequent candidate gene study of one of these cohorts evaluating the different genes encoding the glucocorticoid heterocomplex also identified three SNPs within the heat shock organizing protein gene (STIP1) also associated with lung function response during ICS therapy.18
  | Table 1 Pharmacogenetic candidate genes for inhaled glucocorticoid response in asthma |
The glucocorticoid pathway interacts with other biologic pathways showing gene variation which influences the response to ICS monotherapy or when an ICS is administered with a short-acting beta agonist (SABA) or a long-acting beta agonist (LABA). Adenylyl cyclase type 9 is an enzyme within the canonical β2-adrenergic receptor pathway encoded by ADCY9, a gene with a nonsynonymous or coding SNP, Met772Ile (rs2230739), associated with bronchodilator response to a SABA, albuterol, only in subjects treated with an ICS from the Childhood Asthma Management Program (CAMP) cohort.19 This gene pathway interaction was replicated in an independent Korean trial cohort treated with a LABA, formoterol, in combination with an ICS.20 Another candidate gene for ICS response outside of the glucocorticoid pathway encodes for the T-box expressed in T-cell transcription which regulates naïve T-lymphocyte development (TBX21) and contains a coding SNP, His33Glu (rs2240017), and was associated with improvements in bronchial hyperresponsiveness or bronchoprotection during ICS treatment in the CAMP cohort and replicated in an independent Korean cohort.21,22 A preliminary candidate gene study of CYP3A4, CYP3A5, and CYP3A7 in 413 ICS-treated asthmatic children identified a specific CYP3A4 genotype in 20 children associated with improved asthma symptom control, suggesting that loci important for ICS metabolism (pharmacokinetics) might also serve as biomarkers for therapeutic responsiveness.23
High-throughput genotyping techniques using chip technologies for GWAS have facilitated comprehensive assessment of gene variation for pharmacogenetic studies in asthma cohorts. In GWAS, 500,000 to more than one million SNPs are genotyped to scan the genome in large clinical trial populations. While the number of GWAS in asthma pharmacogenetics is small, the studies performed to date have identified novel pharmacogenetic loci for ICS response. The first GWAS for ICS treatment response was performed in the CAMP cohort (and studied further in 935 asthmatics from four clinical trial cohorts) and identified a promoter SNP in the glucocorticoid-induced transcript-1 gene (rs37972 in GLCCI1) associated with change in lung function during ICS treatment. In vitro, a coinherited promoter variant (rs37973) was transfected into cells and resulted in decreased gene expression.24
This initial asthma pharmacogenetic GWAS was performed in a cohort consisting primarily of children with fewer years of corticosteroid exposure compared with adults, which may, in part, explain why this association was not replicated in a larger clinical trial cohort consisting of adult asthmatics.25 Another GWAS of 408 asthma subjects from the CAMP and the National Heart, Lung, and Blood Institute Childhood Asthma Research and Education trial cohorts also identified SNPs in the T gene (rs3127412 and rs6456042) associated with the lung function response to ICS treatment, which was replicated in an additional 407 ICS-treated asthma subjects.26 GLCCI1 is important for the regulation of apoptosis in response to glucocorticoids, and in combination with other novel loci identified through GWAS, such as the T gene, is an example of novel genetic biomarkers for future genetic profiles.
Biologic candidate gene studies and GWAS have been important in demonstrating that multiple gene variants from interacting pathways influence responsiveness to ICS therapy (Table 1); however, pharmacogenetic loci such as GLCCI1 have accounted for no more than 6.6% of the observed variability in responsiveness to ICS. Thus, additional genetic or other factors in some study cohorts likely account for the variability in ICS responsiveness unaccounted for in current studies. These may be additional gene variants with gene–gene interactions, potentially rare variants not identifiable with GWAS, which in combination determine ICS response.
The potential for multiple loci determining therapeutic responsiveness to ICS is an important rationale for the continued identification of novel loci with additional studies in larger ICS-treated trial cohorts for the development of genetic profiles for personalized approaches. ICS therapy is usually given in combination with other pharmacologic therapies that target additional, potentially interacting pathways; therefore, it is critical to identify biomarkers of responsiveness to these other therapies, which include leukotriene modifiers and inhaled beta agonists.
Cysteinyl leukotriene pathway
While not as therapeutically effective as ICS, leukotriene-modifying drugs are a commonly used adjunct treatment for the management of persistent asthma and are available as two classes of therapies, ie, 5-lipooxygenase (5-LO) inhibitors and cysteinyl leukotriene receptor 1 antagonists. The cysteinyl leukotriene genetic pathway is initiated and limited by 5-LO (encoded by ALOX5) followed by a cascade of enzymes that synthesize different leukotrienes (leukotriene A4 hydrolase [LTA4H] and C4 synthase [LTC4S]) and protein channels (multi-drug resistance protein 1 [MRP1]) that transport leukotrienes to the extracellular space to bind and activate cysteinyl leukotriene receptors (CYSLTR1 and CYSLTR2).
There have been few replicated pharmacogenetic candidate gene studies for leukotriene modifiers in asthma, primarily due to the small sample sizes; however, these studies are informative and demonstrate that the variability in therapeutic responsiveness to these therapies is at least partially determined by gene variation (Table 2). The first of these studies evaluated a tandem repeat polymorphism in the regulatory promoter region of ALOX5 and showed that this promoter variant was associated with lung function response in 114 asthmatics treated with a 5-LO inhibitor, ABT-761.27
  | Table 2 Pharmacogenetic candidate genes for leukotriene modifier response in asthma |
Subsequent larger candidate gene studies have shown that the ALOX5 promoter variant and additional ALOX5 SNPs may also influence the response to the leukotriene receptor antagonist, montelukast.28–31 In addition, variants in LTC4S and MRP1 have been associated with lung function response to another 5-LO inhibitor (zileuton) and montelukast.30,31 These leukotriene pathway candidate gene studies also included SNPs in additional genes, such as LTA4H and CYSLTR1, and have shown conflicting findings likely related to sample sizes (n=61 to n=577) underpowered to consistently detect and replicate pharmacogenetic associations.30,31
In a recent genetic study of the ALOX5 promoter variant in 270 asthmatic children (including 103 non-Hispanic whites and 135 African Americans), homozygotes for the less common repeat alleles (three, four, or six tandem repeats) had a higher urinary concentration of leukotriene E4, consistent with increased leukotriene biosynthesis, and were more likely to be treated with montelukast for asthma control and have lower baseline lung function compared with asthmatics having the more common “wild-type” repeat allele (five tandem repeats). Most importantly, 86% of these minor risk alleles were from African American subjects, suggesting that genetic variation from an African ancestry may influence asthma severity and responsiveness to therapies targeting the leukotriene pathway.32
The variability in responsiveness to leukotriene modifier drugs could relate to interindividual differences in bioavailability influenced by a variant that alters drug pharmacokinetics. The interindividual variability of plasma montelukast levels has been shown to be influenced, at least in part, by carrier-mediated transport in the intestines through an organic anion transporter encoded by the solute carrier organic anion transporter family member 2B1 gene (SLCO2B1). A coding variant in SLCO2B1 (Arg312Gln, rs12422149) was associated with plasma montelukast levels and symptom control during montelukast therapy in a small clinical trial cohort of 80 subjects from different ethnic groups.33 Subsequent smaller studies in controls (n=20 to n=33) were unable to replicate the pharmacokinetic effects of the SLCO2B1 locus on plasma montelukast levels.34,35 Other genes known to be important for the metabolism of 5-LO inhibitors and cysteinyl leukotriene receptor 1 antagonists, such as genes encoding the cytochrome P450 system, have yet to be evaluated.
Pharmacogenetic studies of the leukotriene pathway demonstrate the challenges of performing candidate gene studies in cohorts from clinical trials that were not designed or powered for genetic studies representative of different ethnic groups with varying backgrounds. These studies also show that pharmacokinetic loci could serve as genetic biomarkers for therapeutic responsiveness, including the locus SLCO2B1 which has yet to be replicated in an appropriately powered study cohort (Table 2). Irrespective of these challenges, these studies have identified different loci throughout the cysteinyl leukotriene biosynthetic and receptor pathway that could constitute genetic profiles for personalized therapeutic strategies suited for the small subset of asthma subjects who preferentially respond to these agents compared with ICS therapy.16
β2-adrenergic receptor pathway
Inhaled beta agonists are among the oldest and most commonly prescribed therapies for airways diseases such as asthma and are available in three drug classes: SABA (isoproterenol, fenoterol, levalbuterol, and albuterol or salbutamol), LABA (salmeterol and formoterol), and ultra-long-acting beta agonists (vilanterol and indacaterol). Beta agonists bind the β2-adrenergic receptor to activate a G-protein coupled receptor pathway including adenylyl cyclase type 9 and regulate the production of cyclic AMP and smooth muscle relaxation. These agents have been associated with uncommon asthma-related, life-threatening events since the 1960s and have become the center of a beta agonist controversy that persists today.
Asthma mortality epidemics were associated with a high-dose preparation of a SABA, isoproterenol, in the 1960s, and a potent, less selective SABA, fenoterol, in the 1970s.36–40 These mortality epidemics subsided with the withdrawal of these therapies from the international market. However, a surveillance study in the UK in 1993 followed by SMART (the Salmeterol Asthma Multicenter Research Trial) in the USA in 2005 suggested that LABA treatment could result in a small increase in asthma-related, life-threatening exacerbations or death in asthmatics treated with salmeterol.4,5,36–40 The findings of SMART and subsequent meta-analyses weighed heavily on the SMART cohort formed the basis of a review by the US Food and Drug Administration in 2005 resulting in a public health advisory panel and a boxed warning for all LABAs and LABA-containing combination preparations.41
Subsequent studies have shown that LABA and ICS combination therapy has beneficial effects on asthma control and risk of exacerbations, suggesting that these drugs are safe in the vast majority of asthmatics and that life-threatening adverse responses are likely rare, occurring in a small at-risk subgroup.42,43 However, the most recent US Food and Drug Administration advisory panel in 2010 issued a more detailed boxed warning for LABA-containing preparations and mandated an international LABA safety study in 46,800 asthmatics which is currently underway.41,44 The risk for rare, severe adverse responses to LABA therapy provides a rationale for pharmacogenetic studies to discover genetic biomarkers that could identify the genetic profile of individual asthmatics susceptible to adverse effects. These studies have focused on biologic candidate genes within the β2-adrenergic receptor and nitric oxide synthesis pathways (Table 3).
  | Table 3 Pharmacogenetic candidate genes for β2-adrenergic receptor response in asthma |
The most studied pharmacogenetic locus for beta agonist response is the β2-adrenergic receptor gene (ADRB2), the receptor target for beta agonist therapy. ADRB2 is intronless, yet a polymorphic gene with more than 49 different genetic variants in the multi-ethnic asthma cohorts evaluated to date.45–47 Of these, the most intensively studied is a common coding variant at amino acid position 16, Gly16Arg, which has been associated with altered receptor downregulation in vitro. Early pharmacogenetic studies of ADRB2 demonstrated that Arg16 homozygotes had a greater acute response to SABA bronchodilation compared with Gly16 homozygotes, which has been confirmed in additional asthma populations.48,49
Subsequent pharmacogenetic studies of two independent clinical trial cohorts (on long-term lung function response to regular albuterol treatment) showed that Arg16 homozygotes experienced a decline in peak expiratory flow rate (PEFR), while PEFR remained unchanged in Gly16 homozygotes, prompting one of the first prospective genotype-stratified studies of Gly16Arg genotypes by the National Heart, Lung, and Blood Institute Asthma Clinical Research Network, ie, BARGE (Beta Agonist Response by Genotype).9,50,51 The genotype-stratified design of the BARGE trial ensured that a sufficient number of the less frequent Arg16 homozygotes were randomized to regular albuterol or placebo in this cross-over study (rescue inhaler use was primarily limited to ipratropium to minimize beta agonist exposure). In the BARGE trial, Gly16 homozygotes experienced an increase in PEFR and an improvement in asthma symptom scores and rescue inhaler use during regular albuterol therapy (Figure 2). In contrast, PEFR did not change in Arg16 homozygotes who experienced a deterioration in symptoms and increased rescue inhaler use during regular albuterol treatment (Figure 2).9
  | Figure 2 ACRN Beta Agonist Response by Genotype (BARGE) trial. |
The contrasting effects of the Gly16Arg locus during acute versus long-term SABA therapy could be related to receptor kinetics or inflammatory effects, but do not apply to current asthma treatment guidelines which recommend SABA as a rescue therapy only. Despite this guideline-based recommendation, the practice of regular SABA treatment is still utilized in areas of the world where asthma controller therapies such as LABA and ICS combination preparations are not readily available.52 Pharmacogenetic studies9,48,49,50,51 of the Gly16Arg locus and SABA exposure provided a rationale for studies53,54 of the ADRB2 locus in LABA-treated trial cohorts.
An early pharmacogenetic study of small subgroups from two Asthma Clinical Research Network clinical trials showed that Arg16 homozygotes experienced a decline in PEFR and a deterioration of symptoms during LABA therapy. This adverse response among Arg16 homozygotes was not identified in a pharmacogenetic study of a cross-over trial of SABA and LABA treatment, nor was it confirmed in subsequent studies of an additional three clinical trial cohorts, including a substantially larger cohort of nearly 2,000 asthmatics. In addition, two subsequent prospective, genotype-stratified clinical trials were not able to identify significant differences in PEFR responsiveness to LABA therapy between Gly16Arg genotypes.53,54
A common ADRB2 variant, such as Gly16Arg, has an allele frequency of 40%–50% in ethnic groups from the USA; however, other racial groups may have a higher frequency of the Arg16 allele, including Asian populations. Thus, Gly16Arg likely has a small effect on LABA responsiveness and it is more likely that a rare variant with a strong biologic effect could account for the uncommon, severe adverse responses observed in less than 1% of the subjects randomized to LABA in the SMART cohort (Figure 3). Resequencing of ADRB2 in multi-ethnic asthma populations has identified different rare variants with a frequency of less than 5%, most of which were unique to a particular ethnic group.45,46 For instance, Thr164Ile is a rare coding variant, more common in non-Hispanic whites, while a 24-base-pair promoter insertion variant at nucleotide position -376 relative to the start codon (ie, -376 In-Del) was only identified in African Americans and Puerto Ricans.45–47
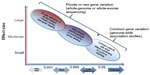  | Figure 3 Impact of genetic variants in human disease. |
Thr164Ile is located within the fourth transmembrane domain of the receptor and results in a marked reduction in Gs-protein signaling and ligand binding affinity in response to different SABAs and LABAs in vitro.55,56 The Ile164 variant also impairs binding of salmeterol to the receptor’s “salmeterol exosite”.56 In nearly 60,000 subjects from two Copenhagen-based general population studies that included a subgroup of 1,300 self-reported asthmatics, this rare variant was also associated with impaired baseline lung function and risk of airflow obstruction.57
In a recent pharmacogenetic study of rare ADRB2 variants, the rare Thr164Ile and -376 In-Del variants were associated with asthma-related hospitalization in the past year in non-Hispanic white and African American asthmatics treated with a LABA, respectively (Figure 4A and B). Each rare ADRB2 variant was also associated with asthma-related urgent outpatient visits and a requirement for regular systemic glucocorticoid use in LABA-treated subjects from each ethnic group. This increased risk of severe exacerbations and poor symptom control was only observed in LABA-treated asthmatic subjects in each ethnic group, suggesting a gene-by-environment interaction for these two rare ADRB2 variants (Figure 4A and B).46 These rare variants are an example of the importance of considering different rare variants unique to different ethnic groups in pharmacogenetic studies. Thus, rare variants such as those in ADRB2 are potential biomarkers for more personalized, precise, guideline-based treatment strategies in the small subgroup of asthmatics with altered responsiveness to LABA and ICS combination therapy.
  | Figure 4 Two rare ADRB2 variants and asthma-related hospitalization with LABA treatment. |
Biologic candidate gene studies of beta agonist response have also included genes within the G-protein coupled β2-adrenergic receptor pathway. ADCY9, a canonical receptor pathway gene, has a coding variant, Ile772Met, associated with acute bronchodilation in response to SABA in ICS-treated asthmatics from the CAMP study and lung function response to a LABA and ICS treatment in a Korean asthma population.19,20 The corticotropin-releasing hormone receptor-2 is a G-coupled protein receptor that regulates relaxation of smooth muscle via activation of adenylyl cyclase and protein kinase A encoded by CRHR2. Five SNPs in CRHR2 have been associated with an acute SABA bronchodilator response in three independent cohorts.58
Recent studies have also shown that rare variants adjacent to ADCY9 and CRHR2 are associated with albuterol bronchodilator response in Puerto Rican and Mexican asthma subjects from the GALA (Genetics in Latino Americans) cohort.59 These findings suggest that gene variation within a G-protein coupled receptor pathway influence bronchodilator response to SABAs; however, studies need to be performed in clinical trial cohorts treated with a LABA.
Arginase-1 and arginase-2 metabolize L-arginine, a natural substrate for nitric oxide synthase, to generate nitric oxide, which is an endogenous bronchodilator. The genes that code for these enzymes (ARG1 and ARG2) have been the focus of biologic candidate genes within the nitric oxide biosynthetic pathway. Different ARG1 variants were associated with an acute SABA bronchodilator response in asthmatics from CAMP and have been replicated in independent asthma trial cohorts, while ARG2 SNPs have also been associated with SABA response in a Dutch asthma cohort.60–62 In addition, a coding variant in the endothelial nitric oxide synthase gene (Asp298Glu in NOS3) was associated with lung function response to LABA and ICS combination therapy in a small study of 81 asthmatic children.63
Variation within pathway-related genes may interact with each other or with other pathways to predict responsiveness to beta agonists in the context of a genetic profile. Genome-wide approaches have the potential to identify novel interacting loci. A GWAS was performed in 1,644 non-Hispanic white asthmatics from six different clinical trial cohorts and identified a promoter SNP in SPATS2L (rs295137) associated with an acute albuterol bronchodilator response. In human airway smooth muscle cells, miRNA knockdown of SPATS2L resulted in increased β2-adrenergic receptor expression, confirming its role in this therapeutic receptor pathway.64 A recent GWAS of 403 non-Hispanic whites from CAMP with two-tiered replication in four asthma cohorts identified an intergenic SNP (rs11252394) on chromosome 10p15 associated with SABA response. This SNP was adjacent to potential biologic candidate genes, such as that encoding for protein kinase C (PRKCQ); however, more detailed genotyping (ie, fine mapping) of this genomic region and replication will be necessary to evaluate whether this SNP is coinherited with a functional variant.65
Pharmacogenetic studies and biologic drug development
The development of novel biologic therapies targeting different biologic pathways in asthma is currently in progress and holds the promise of advancing personalized medicine. The interleukin (IL)-4 and IL-13 pathway mediates Th2 lymphocyte-mediated allergic inflammation by binding and activating a common subunit of the IL-4 receptor, the IL-4α receptor subunit. In recent clinical trials, a molecular inhibitor of the IL-4α receptor subunit, pitrakinra, and a monoclonal antibody, dupilumab, have been shown to be effective in preventing loss of symptom control in asthma subpopulations characterized by increased blood or sputum eosinophils. Both biologic drugs block the IL-4α receptor subunit (encoded by IL4RA), resulting in dual inhibition of a shared IL-4 and IL-13 proinflammatory pathway.66,67
In a dose-ranging study of pitrakinra, the primary endpoint of a study-defined exacerbation was significantly reduced, with a significant dose-response in asthmatics with a specific IL4RA variant genotype (GG of rs8832, nearly one-third of the cohort) while no dose response was observed in subjects with the remaining genotypes (AG or AA at rs8832).67 The overall findings of the pitrakinra clinical trial were negative; however, the study did not recruit based on markers for eosinophilic inflammation or IL4RA genotype. Most importantly, this clinical trial was an example of how a pharmacogenetic biomarker can identify a subgroup of responders embedded within an overall cohort which was unresponsive to a biologic drug. Targeted biologic therapies for asthma will inevitably be expensive and should only be provided to a subgroup with definitive biomarker evidence of responsiveness, possibly based on a pharmacogenetic parameter.
Novel pharmacogenetic approaches
One of the most important challenges of pharmacogenetics research in asthma is that therapeutic responsiveness in a complex disease of multiple genetic and environmental factors is likely influenced by a large number of different pharmacogenetic loci in interacting pathways. This challenge has become evident as different pharmacogenetic studies24,64 have made discoveries that only account for a small proportion of the variability in drug responsiveness or have been difficult to replicate in independent study cohorts. For many candidate gene studies, inconsistencies in replication of biologic candidate genes may have been due to underpowered sample sizes, or differences in genetic ancestry or population structure between study cohorts that alter the frequency of associated variants, gene–gene interactions, and environmental factors.68 An important factor that could influence therapeutic responsiveness in asthma includes gene variation, which varies in frequency between different ancestral populations, such as low-frequency or rare variants, and could be unique pharmacogenetic loci for an individual racial or ethnic group.
Whole-genome sequencing data from the 1,000 Genomes Project Consortium recently identified an enrichment of rare variants, and found that individual genomes from an African ancestry had a three times greater frequency of rare variants compared with other ancestries.69 The allele frequencies of variants throughout the genome, particularly less common or rare variants, vary in frequency between different ethnic or racial groups, and thus could potentially be identified as pharmacogenetic loci using mapping by admixture linkage or admixture mapping.68 Admixture mapping takes advantage of the variable allele frequencies of SNPs between recently admixed ethnic groups to test for associations between ancestral estimates at a particular SNP with a phenotype such as drug response. Admixture mapping has recently been used in conjunction with GWAS in 1,782 Puerto Rican and Mexican children from the GALA cohort to identify novel rare variants within two solute carrier genes associated with a SABA bronchodilator response.59 It will be important to consider rare variants in future pharmacogenetic studies since these variants are being increasingly recognized as loci that may influence therapeutic responsiveness and contribute to more individualized precise predictions for personalized approaches.5,68
Additional factors that could influence therapeutic responsiveness in asthma include interactions between variants from different genes (gene–gene interactions) and gene–environment interactions. Coincident polymorphisms in different genes or within the same gene as a haplotype can result in interactions that magnify or diminish pharmacogenetic effects.70 In Puerto Ricans with asthma, the Gly16Arg variant in ADRB2 has been shown to interact with variation in S-nitrosoglutathione reductase to additively influence the bronchodilator response to albuterol.71 Gene–gene interactions are fundamental to the development of genetic profiles for personalized therapeutic approaches because the required panels of genetic biomarkers will likely encompass variants from different genes which interact additively in a more complex manner to influence therapeutic responsiveness. Systems biology approaches can take advantage of large datasets from whole-genome genotyping to identify novel pharmacogenetic gene–gene interactions. For instance, such an approach identified novel gene–gene interactions in 308 non-Hispanic white asthmatics from CAMP, which consisted of a network of 15 SNPs within 15 different genes that predicted SABA bronchodilation more accurately than each individual locus.72
In vitro studies have also identified novel pharmacogenetic loci that interact with known therapeutic pathways through differential gene expression. For instance, the thyroid hormone receptor-β gene (THRB) was found to be differentially expressed in response to beta agonist exposure in vitro and contained a promoter SNP associated with SABA bronchodilator response in asthmatics from CAMP and three clinical trial cohorts.73 In addition to regulatory genetic variants (SNPs), epigenetic modifications (ie, DNA methylation) regulate gene expression in response to environmental exposures and have been implicated in asthma susceptibility and severity in a small number of studies.74–78 The promoter region of ADRB2 has been shown to undergo differential methylation that was associated with asthma severity in small pediatric cohorts; however, there is a paucity of pharmacogenetic studies evaluating epigenetic variation in asthma.77,78
Pharmacogenetic discoveries and development of genetic profiles
Personalized medicine is a new model of health care that delivers preventative or therapeutic options based on the unique characteristics of an individual patient. This review has focused on genetic biomarkers from pharmacogenetic studies predictive of therapeutic responsiveness to commonly used pharmacologic therapies; however, demographic, environmental, or inflammatory biomarkers from peripheral blood and airway samples could also provide information about responsive subgroups. Therefore, we have also focused on how pharmacogenetic research has identified different biomarkers for genetic profiles that could be used to tailor therapy for an individual asthmatic, but genetic loci for asthma susceptibility or risk variants could also be used to decide which unaffected individuals would benefit from preventative intervention.
The ideal pharmacogenetic profile for personalized precision medicine will likely consist of a few key features. First, the ideal genetic profile would include a panel of common and rare variants predictive of therapeutic responsiveness to different treatment options based on an individual’s ancestral background. Genetic tests or panels have been developed for both rare and common diseases; however, the most predictive biomarkers for therapeutic response in asthma have been based on single variants.13,79,80 Since therapeutic responsiveness in asthma likely results from multiple genes and gene–gene interactions, these single-variant biomarkers have had low predictive accuracy.6 Thus, genetic profile panels with multiple associated variants should be developed, validated in new cohorts, and updated as new pharmacogenetic discoveries emerge.81
The existing catalog of genetic variation has continued to expand as the costs associated with whole-genome genotyping and next-generation sequencing have come down. For instance, large-scale whole-exome and whole-genome sequencing projects such as the National Institutes of Health National Heart, Lung, and Blood Institute GO Exome Sequencing Program, the 1,000 Genomes Project, and the Consortium on Asthma in African Ancestry Populations have resulted in an expanding database of common and rare genetic variants for future pharmacogenetic studies in different racial and ethnic groups.68,69
Second, pharmacogenetic profiles of the future will contain gene variants predictive of alternative phenotypes representing adverse responses, which may vary between individuals from different ancestral backgrounds. These genetic biomarkers would prevent adverse events that could be life-threatening. Detailed sequencing of ADRB2 in multi-ethnic asthma populations identified a gene-by-environment interaction for two different rare ADRB2 variants and the risk for hospitalization during LABA treatment in two different ethnic groups.46 These rare variant effects on adverse LABA responses requires replication in a large trial cohort, such as the LABA-mandated safety study cohort of 46,800 asthma subjects, but emphasize the impact of detailed sequencing in populations from different ancestral backgrounds to identify uncommon pharmacogenetic loci with a strong impact on drug response.44,46 In the management of severe asthmatics who remain symptomatic despite multiple therapies, these rare variants are an example of a potential biomarker that could determine whether discontinuation of the LABA would have beneficial effects on symptom control or whether an alternative therapy such as a long-acting muscarinic drug or a biologic drug may be the most efficacious treatment option.
Third, the ideal pharmacogenetic profile should account for gene–gene interactions in different therapeutic biologic pathways, particularly in the setting of combination therapy with different pharmacologic therapies such as LABA and ICS. Such a panel would be predictive of the best initial course of therapy but also predict the most efficacious adjunctive therapeutic option for an individual asthmatic receiving a first-line therapy such as an ICS. For instance, when a LABA and ICS are used in combination, it is possible that genetic variants in the β2-adrenergic receptor pathway result in a beneficial therapeutic effect yet interact with other variants in the glucocorticoid pathway. The resulting gene–gene interactions could magnify, attenuate, or neutralize clinical responses during LABA and ICS combination therapy. The variability in allele frequencies between different racial or recently admixed ethnic groups also has the potential to alter these gene–gene interactions and therapeutic responsiveness.68
Fourth, pharmacogenetic profiles of the future should also contain loci shown to alter drug metabolism or pharmacokinetic parameters and impact bioavailability. These pharmacogenetic biomarkers would be important for drug dosing in an individual asthmatic and be predictive of the optimal dose of a pharmacologic therapy to achieve maximum beneficial effects at a minimal risk for toxicity from supratherapeutic dosing. Preliminary studies have identified CYP3A4 and SLCO2B1 as loci that may alter drug pharmacokinetics to influence responsiveness to ICS and montelukast therapy.23,33 These findings require replication but illustrate the potential for pharmacokinetic loci as biomarkers for therapeutic responsiveness to different pharmacologic therapies in asthma.
Fifth, the ideal pharmacogenetic profile should account for the appropriate response phenotypes in the appropriate subgroup of patients. For instance, severe asthma exacerbations requiring hospitalization (a primary outcome for the safety study41,44 mandated by the US Food and Drug Administration) might be a more accurate surrogate outcome for rare, life-threatening adverse responses to LABA therapy compared with more common phenotypes, such as lung function responsiveness.In addition, a subgroup of more severe asthmatics with a history of frequent asthma-related health care utilization might be a more appropriate cohort to evaluate LABA safety, but these patients are excluded from most clinical trials.5,44,46 Similarly, a pharmacogenetic study limited to the responder subgroup from a clinical trial cohort treated with montelukast resulted in a panel of four genetic variants within the cysteinyl leukotriene receptor pathway which accounted for a greater proportion of the variability in therapeutic responsiveness compared with a combined analysis with nonresponders.82 Identification of an appropriate subgroup of responders has been critical to the development of novel biologic drugs and will be important to consider for all classes of asthma therapies in future pharmacogenetic studies and the development of genetic profiles.66,67
Conclusion
Personalized medicine places the individual at the center of health care through individual characteristics such as genetic predictive biomarkers.6 Panels of genes and other biomarkers are being developed to individualize treatment for asthmatics, particularly when using expensive biologic drugs in those with severe disease unresponsive to a combination of different pharmacologic therapies. Genetic profiles in this setting would be based on pharmacogenetic biomarkers that would tailor treatment strategies based on the predominant molecular pathways for an individual asthmatic who could then be treated with a specific pharmacologic therapy or combination of therapies.6 The benefit of this personalized, tailored approach is most evident in the development of expensive biologic drugs, where a pathway-directed genetic profile could minimize unnecessary or prolonged exposure to these expensive therapies in those less likely to be responsive.
Therapeutic responsiveness to more commonly used therapies is likely determined by multiple gene variants. Thus, genetic profiles based on pharmacogenetic loci identified to date would still have a low accuracy in predicting individual responsiveness to a specific therapy. Pharmacogenetic approaches are rapidly expanding and it is possible that current discoveries in combination with additional variants identified in future studies will form the basis of genetic profiles for personalized, tailored approaches to more commonly used therapies.
Pharmacogenetic approaches have already become useful in other pulmonary diseases to identify individuals most likely to respond to costly biologic therapies. In cystic fibrosis, a rare, functional genetic variant in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR), Gly551Asp, identified a subgroup of patients who experienced substantial benefits from ivacaftor, a pharmacologic agent targeting the resulting channel gating abnormality.13 Recently, a Phase III clinical trial showed that those with eight additional rare CFTR coding variants also experienced beneficial effects in response to ivacaftor. It is possible that variants in candidate genes such as IL4RA could also serve as biomarkers for biologic drugs currently under development, such as dupilumab.66,67
Future advances in pharmacogenetics research will depend on a continued collaborative effort in order to recruit and analyze larger, comprehensively characterized asthma populations from different racial and ethnic groups representative of different ancestral backgrounds. In addition, DNA should be aggressively collected in all drug trial cohorts and made available for pharmacogenetic studies. In time, the costs of high-throughput genotyping and DNA sequencing will continue to decrease, while the expanding volume of genetic data will be deciphered by different analytic methods that take into account genetic ancestry, gene–gene interactions, and rare variant effects. An integrated approach will be critical for the development of genetic profiles based on the complex molecular mechanisms underlying treatment responses to eventually deliver truly precise and personalized medicine to an individual asthmatic.
Acknowledgment
This work is supported by the National Institutes of Health National Heart, Lung, and Blood Institute (grants R01 NR013700, U10 HL109164, U01 HL 65899, and U10 HL098103).
Disclosure
The authors report no conflicts of interest in this work.
References
[No authors listed]. Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. American Thoracic Society. Am J Respir Crit Care Med. 2000;162(6):2341–2351. | |
Chan MT, Leung DY, Szefler SJ, Spahn JD. Difficult-to-control asthma: clinical characteristics of steroid-insensitive asthma. J Allergy Clin Immunol. 1998;101(5):594–601. | |
Salpeter SR, Buckley NS, Ormiston TM, Salpeter EE. Meta-analysis: effect of long-acting beta-agonists on severe asthma exacerbations and asthma-related deaths. Ann Intern Med. 2006;144(12):904–912. | |
Castle W, Fuller R, Hall J, Palmer J. Serevent nationwide surveillance study: comparison of salmeterol with salbutamol in asthmatic patients who require regular bronchodilator treatment. BMJ. 1993;306(6884):1034–1037. | |
Nelson HS, Weiss ST, Bleecker ER, Yancey SW, Dorinsky PM. The Salmeterol Multicenter Asthma Research Trial: a comparison of usual pharmacotherapy for asthma or usual pharmacotherapy plus salmeterol. Chest. 2006;129(1):15–26. | |
Meyers DA, Bleecker ER, Holloway JW, Holgate ST. Asthma genetics and personalised medicine. Lancet Respir Med. 2014;2(5):405–415. | |
Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56(4):1054–1070. | |
Kalow W, Tang BK, Endrenyi L. Hypothesis: comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics. 1998;8(4):283–289. | |
Israel E, Chinchilli VM, Ford JG, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004;364(9444):1505–1512. | |
Li X, Hawkins GA, Ampleford EJ, et al. Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. J Allergy Clin Immunol. 2013;132(2):313–320. e315. | |
Li X, Howard TD, Moore WC, et al. Importance of hedgehog interacting protein and other lung function genes in asthma. J Allergy Clin Immunol. 2011;127(6):1457–1465. | |
Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382(9893):720–731. | |
Ramsey BW, Davies J, McElvaney NG, et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N Engl J Med. 2011;365(18):1663–1672. | |
Peters SP, Anthonisen N, Castro M, et al. Randomized comparison of strategies for reducing treatment in mild persistent asthma. N Engl J Med. 2007;356(20):2027–2039. | |
Sorkness CA, Lemanske RF Jr, Mauger DT, et al. Long-term comparison of 3 controller regimens for mild-moderate persistent childhood asthma: the Pediatric Asthma Controller Trial. J Allergy Clin Immunol. 2007;119(1):64–72. | |
Szefler SJ, Phillips BR, Martinez FD, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol. 2005;115(2):233–242. | |
Tantisira KG, Lake S, Silverman ES, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13(13):1353–1359. | |
Hawkins GA, Lazarus R, Smith RS, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123(6):1376–1383. e1377. | |
Tantisira KG, Small KM, Litonjua AA, Weiss ST, Liggett SB. Molecular properties and pharmacogenetics of a polymorphism of adenylyl cyclase type 9 in asthma: interaction between beta-agonist and corticosteroid pathways. Hum Mol Genet. 2005;14(12):1671–1677. | |
Kim SH, Ye YM, Lee HY, Sin HJ, Park HS. Combined pharmacogenetic effect of ADCY9 and ADRB2 gene polymorphisms on the bronchodilator response to inhaled combination therapy. J Clin Pharm Ther. 2011;36(3):399–405. | |
Tantisira KG, Hwang ES, Raby BA, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci U S A. 2004;101(52):18099–18104. | |
Ye YM, Lee HY, Kim SH, et al. Pharmacogenetic study of the effects of NK2R G231E G>A and TBX21 H33Q C>G polymorphisms on asthma control with inhaled corticosteroid treatment. J Clin Pharm Ther. 2009;34(6):693–701. | |
Stockmann C, Fassl B, Gaedigk R, et al. Fluticasone propionate pharmacogenetics: CYP3A4*22 polymorphism and pediatric asthma control. J Pediatr. 2013;162(6):1222–1227. e1–e2. | |
Tantisira KG, Lasky-Su J, Harada M, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365(13):1173–1183. | |
Hosking L, Bleecker E, Ghosh S, et al. GLCCI1 rs37973 does not influence treatment response to inhaled corticosteroids in white subjects with asthma. J Allergy Clin Immunol. 2014;133(2):587–589. | |
Tantisira KG, Damask A, Szefler SJ, et al. Genome-wide association identifies the T gene as a novel asthma pharmacogenetic locus. Am J Respir Crit Care Med. 2012;185(12):1286–1291. | |
Drazen JM, Yandava CN, Dube L, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat Genet. 1999;22(2):168–170. | |
Telleria JJ, Blanco-Quiros A, Varillas D, et al. ALOX5 promoter genotype and response to montelukast in moderate persistent asthma. Respir Med. 2008;102(6):857–861. | |
Klotsman M, York TP, Pillai SG, et al. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet Genomics. 2007;17(3):189–196. | |
Lima JJ, Zhang S, Grant A, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006;173(4):379–385. | |
Tantisira KG, Lima J, Sylvia J, Klanderman B, Weiss ST. 5-lipoxygenase pharmacogenetics in asthma: overlap with Cys-leukotriene receptor antagonist loci. Pharmacogenet Genomics. 2009;19(3):244–247. | |
Mougey E, Lang JE, Allayee H, et al. ALOX5 polymorphism associates with increased leukotriene production and reduced lung function and asthma control in children with poorly controlled asthma. Clin Exp Allergy. 2013;43(5):512–520. | |
Mougey EB, Feng H, Castro M, Irvin CG, Lima JJ. Absorption of montelukast is transporter mediated: a common variant of OATP2B1 is associated with reduced plasma concentrations and poor response. Pharmacogenet Genomics. 2009;19(2):129–138. | |
Kim KA, Lee HM, Joo HJ, Park IB, Park JY. Effects of polymorphisms of the SLCO2B1 transporter gene on the pharmacokinetics of montelukast in humans. J Clin Pharmacol. 2013;53(11):1186–1193. | |
Tapaninen T, Karonen T, Backman JT, Neuvonen PJ, Niemi M. SLCO2B1 c.935G>A single nucleotide polymorphism has no effect on the pharmacokinetics of montelukast and aliskiren. Pharmacogenet Genomics. 2013;23(1):19–24. | |
Grainger J, Woodman K, Pearce N, et al. Prescribed fenoterol and death from asthma in New Zealand, 1981-7: a further case-control study. Thorax. 1991;46(2):105–111. | |
Crane J, Pearce N, Burgess C, Jackson R, Beasley R. End of New Zealand asthma epidemic. Lancet. 1995;345(8955):984–985. | |
Pearce N, Grainger J, Atkinson M, et al. Case-control study of prescribed fenoterol and death from asthma in New Zealand, 1977-81. Thorax. 1990;45(3):170–175. | |
Pearce N, Burgess C, Crane J, Beasley R. Fenoterol, asthma deaths, and asthma severity. Chest. 1997;112(4):1148–1150. | |
Stolley PD. Asthma mortality. Why the United States was spared an epidemic of deaths due to asthma. Am Rev Respir Dis. 1972;105(6):883–890. | |
US Food and Drug Administration. FDA Drug Safety Communication: FDA requires post-market safety trials for long-acting beta-agonists (LABAs). 2011. Available from: http://www.fda.gov/Drugs/DrugSafety/ucm251512.htm. Accessed July 10, 2012. | |
Peters SP, Prenner BM, Mezzanotte WS, Martin P, O’Brien CD. Long-term safety and asthma control with budesonide/formoterol versus budesonide pressurized metered-dose inhaler in asthma patients. Allergy Asthma Proc. 2008;29(5):499–516. | |
Sears MR, Ottosson A, Radner F, Suissa S. Long-acting beta-agonists: a review of formoterol safety data from asthma clinical trials. Eur Respir J. 2009;33(1):21–32. | |
Chowdhury BA, Seymour SM, Levenson MS. Assessing the safety of adding LABAs to inhaled corticosteroids for treating asthma. N Engl J Med. 2011;364(26):2473–2475. | |
Hawkins GA, Tantisira K, Meyers DA, et al. Sequence, haplotype, and association analysis of ADRbeta2 in a multiethnic asthma case-control study. Am J Respir Crit Care Med. 2006;174(10):1101–1109. | |
Ortega VE, Hawkins GA, Moore WC, et al. Effect of rare genetic variants in ADRB2 on risk of severe exacerbations and symptom control during longacting beta agonist treatment in a multiethnic asthma population. Lancet Respir Med. 2014;2(3):204–213. | |
Drysdale CM, McGraw DW, Stack CB, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci U S A. 2000;97(19):10483–10488. | |
Choudhry S, Ung N, Avila PC, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am J Respir Crit Care Med. 2005;171(6):563–570. | |
Lima JJ, Thomason DB, Mohamed MH, Eberle LV, Self TH, Johnson JA. Impact of genetic polymorphisms of the beta2-adrenergic receptor on albuterol bronchodilator pharmacodynamics. Clin Pharmacol Ther. 1999;65(5):519–525. | |
Taylor DR, Drazen JM, Herbison GP, Yandava CN, Hancox RJ, Town GI. Asthma exacerbations during long term beta agonist use: influence of beta(2) adrenoceptor polymorphism. Thorax. 2000;55(9):762–767. | |
Israel E, Drazen JM, Liggett SB, et al. The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000;162(1):75–80. | |
Bousquet J, Bousquet PJ, Godard P, Daures JP. The public health implications of asthma. Bull World Health Organ. 2005;83(7):548–554. | |
Wechsler ME, Kunselman SJ, Chinchilli VM, et al. Effect of beta2-adrenergic receptor polymorphism on response to longacting beta2 agonist in asthma (LARGE trial):a genotype-stratified, randomised, placebo-controlled, crossover trial. Lancet. 2009;374(9703):1754–1764. | |
Bleecker ER, Nelson HS, Kraft M, et al. Beta2-receptor polymorphisms in patients receiving salmeterol with or without fluticasone propionate. Am J Respir Crit Care Med. 2010;181(7):676–687. | |
Green SA, Cole G, Jacinto M, Innis M, Liggett SB. A polymorphism of the human beta 2-adrenergic receptor within the fourth transmembrane domain alters ligand binding and functional properties of the receptor. J Biol Chem. 1993;268(31):23116–23121. | |
Green SA, Rathz DA, Schuster AJ, Liggett SB. The Ile164 beta(2)-adrenoceptor polymorphism alters salmeterol exosite binding and conventional agonist coupling to G(s). Eur J Pharmacol. 2001;421(3):141–147. | |
Thomsen M, Nordestgaard BG, Sethi AA, Tybjaerg-Hansen A, Dahl M. beta2-adrenergic receptor polymorphisms, asthma and COPD: two large population-based studies. Eur Respir J. 2012;39(3):558–566. | |
Poon AH, Tantisira KG, Litonjua AA, et al. Association of corticotropin-releasing hormone receptor-2 genetic variants with acute bronchodilator response in asthma. Pharmacogenet Genomics. 2008;18(5):373–382. | |
Drake KA, Torgerson DG, Gignoux CR, et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J Allergy Clin Immunol. 2014;133(2):370–378. | |
Litonjua AA, Lasky-Su J, Schneiter K, et al. ARG1 is a novel bronchodilator response gene: screening and replication in four asthma cohorts. Am J Respir Crit Care Med. 2008;178(7):688–694. | |
Vonk JM, Postma DS, Maarsingh H, Bruinenberg M, Koppelman GH, Meurs H. Arginase 1 and arginase 2 variations associate with asthma, asthma severity and beta2 agonist and steroid response. Pharmacogenet Genomics. 2010;20(3):179–186. | |
Duan QL, Gaume BR, Hawkins GA, et al. Regulatory haplotypes in ARG1 are associated with altered bronchodilator response. Am J Respir Crit Care Med. 2011;183(4):449–454. | |
Iordanidou M, Paraskakis E, Tavridou A, Paschou P, Chatzimichael A, Manolopoulos VG. G894T polymorphism of eNOS gene is a predictor of response to combination of inhaled corticosteroids with long-lasting beta2-agonists in asthmatic children. Pharmacogenomics. 2012;13(12):1363–1372. | |
Himes BE, Jiang X, Hu R, et al. Genome-wide association analysis in asthma subjects identifies SPATS2L as a novel bronchodilator response gene. PLoS Genet. 2012;8(7):e1002824. | |
Duan QL, Lasky-Su J, Himes BE, et al. A genome-wide association study of bronchodilator response in asthmatics. Pharmacogenomics J. 2014;14(1):41–47. | |
Wenzel S, Ford L, Pearlman D, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med. 2013;368(26):2455–2466. | |
Slager RE, Otulana BA, Hawkins GA, et al. IL-4 receptor polymorphisms predict reduction in asthma exacerbations during response to an anti-IL-4 receptor alpha antagonist. J Allergy Clin Immunol. 2012;130(2):516–522. e514. | |
Ortega VE, Meyers DA. Pharmacogenetics: Implications of race and ethnicity on defining genetic profiles for personalized medicine. J Allergy Clin Immunol. 2014;133(1):16–26. | |
1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. | |
Ortega VE, Wechsler ME. Asthma pharmacogenetics: responding to the call for a personalized approach. Curr Opin Allergy Clin Immunol. 2013;13(4):399–409. | |
Choudhry S, Que LG, Yang Z, et al. GSNO reductase and beta2-adrenergic receptor gene-gene interaction: bronchodilator responsiveness to albuterol. Pharmacogenet Genomics. 2010;20(6):351–358. | |
Himes BE, Wu AC, Duan QL, et al. Predicting response to short-acting bronchodilator medication using Bayesian networks. Pharmacogenomics. 2009;10(9):1393–1412. | |
Duan QL, Du R, Lasky-Su J, et al. A polymorphism in the thyroid hormone receptor gene is associated with bronchodilator response in asthmatics. Pharmacogenomics J. 2013;13(2):130–136. | |
Begin P, Nadeau KC. Epigenetic regulation of asthma and allergic disease. Allergy Asthma Clin Immunol. 2014;10(1):27. | |
Michel S, Busato F, Genuneit J, et al. Farm exposure and time trends in early childhood may influence DNA methylation in genes related to asthma and allergy. Allergy. 2013;68(3):355–364. | |
Soto-Ramirez N, Arshad SH, Holloway JW, et al. The interaction of genetic variants and DNA methylation of the interleukin-4 receptor gene increase the risk of asthma at age 18 years. Clin Epigenetics. 2013;5(1):1. | |
Gaffin JM, Raby BA, Petty CR, et al. beta-2 adrenergic receptor gene methylation is associated with decreased asthma severity in inner-city schoolchildren: asthma and rhinitis. Clin Exp Allergy. 2014;44(5):681–689. | |
Fu A, Leaderer BP, Gent JF, Leaderer D, Zhu Y. An environmental epigenetic study of ADRB2 5'-UTR methylation and childhood asthma severity. Clin Exp Allergy. 2012;42(11):1575–1581. | |
Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358(9):910–919. | |
Thanassoulis G, Peloso GM, Pencina MJ, et al. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5(1):113–121. | |
Aronson SJ, Clark EH, Varugheese M, Baxter S, Babb LJ, Rehm HL. Communicating new knowledge on previously reported genetic variants. Genet Med. April 5, 2012. [Epub ahead of print.] | |
York TP, Vargas-Irwin C, Anderson WH, van den Oord EJ. Asthma pharmacogenetic study using finite mixture models to handle drug-response heterogeneity. Pharmacogenomics. 2009;10(5):753–767. | |
Ortega VE, Hawkins GA, Peters SP, Bleecker ER. Pharmacogenetics of the beta 2-adrenergic receptor gene. Immunol Allergy Clin North Am. 2007;27(4):665–684; vii. | |
Tsuji S. Genetics of neurodegenerative diseases: insights from high-throughput resequencing. Hum Mol Genet. 2010;19(R1):R65–R70. | |
Martinez FD, Graves PE, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J Clin Invest. 1997;100(12):3184–3188. | |
Wechsler ME, Lehman E, Lazarus SC, et al. Beta-adrenergic receptor polymorphisms and response to salmeterol. Am J Respir Crit Care Med. 2006;173(5):519–526. | |
Bleecker ER, Postma DS, Lawrance RM, Meyers DA, Ambrose HJ, Goldman M. Effect of ADRB2 polymorphisms on response to longacting beta2-agonist therapy: a pharmacogenetic analysis of two randomised studies. Lancet. 2007;370(9605):2118–2125. | |
Bleecker ER, Yancey SW, Baitinger LA, et al. Salmeterol response is not affected by beta2-adrenergic receptor genotype in subjects with persistent asthma. J Allergy Clin Immunol. 2006;118(4):809–816. | |
Lee DK, Currie GP, Hall IP, Lima JJ, Lipworth BJ. The arginine-16 beta2-adrenoceptor polymorphism predisposes to bronchoprotective subsensitivity in patients treated with formoterol and salmeterol. Br J Clin Pharmacol. 2004;57(1):68–75. | |
Lipworth BJ, Basu K, Donald HP, et al. Tailored second-line therapy in asthmatic children with the Arg16 genotype. Clin Sci (Lond). 2013;124(8):521–528. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
