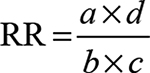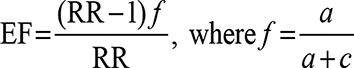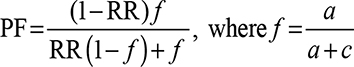Back to Journals » Journal of Inflammation Research » Volume 9
Association of tumor necrosis factor-α and -β gene polymorphisms in inflammatory bowel disease
Authors Al-Meghaiseeb E, Al-Robayan A, Al-Otaibi M, Arfin M, Al-Asmari A
Received 24 November 2015
Accepted for publication 16 January 2016
Published 17 June 2016 Volume 2016:9 Pages 133—140
DOI https://doi.org/10.2147/JIR.S101225
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Ebtissam Saleh Al-Meghaiseeb,1 Abdulrahman A Al-Robayan,1 Mulfi Mubarak Al-Otaibi,1 Misbahul Arfin,2 Abdulrahman K Al-Asmari2
1Department of Gastroenterology, 2Research Centre, Prince Sultan Military Medical City, Riyadh, Saudi Arabia
Abstract: Inflammatory bowel disease (IBD) is a complex, multifactorial, chronic inflammatory disorder of the gastrointestinal tract in which immune dysregulation caused by genetic and/or environmental factors plays an important role. The aim of this case–control study was to evaluate the association of tumor necrosis factor-alpha (TNF-α) (308) and -β (+252) polymorphisms with susceptibility of IBD. A total of 379 Saudi subjects including 179 IBD patients (ulcerative colitis (UC) =84 and Crohn’s disease (CD) =95) and 200 age- and sex-matched healthy controls were recruited. TNF-a and TNF-b genes were amplified using an amplification refractory mutation systems polymerase chain reaction methodology to detect TNF-α (–308) and -β (+252) polymorphisms. The frequency of the GA genotype of TNF-α (–308G/A) was higher, and the frequencies of the GG and AA genotypes were significantly lower in IBD patients compared with those in controls, indicating that genotype GA-positive individuals are susceptible to IBD and that the GG and AA genotypes exert a protective effect. The frequency of allele A of TNF-α (–308G/A) was significantly higher and that of allele G was lower in IBD patients compared with those in controls, indicating an association of allele A with IBD risk in Saudi patients. On stratification of IBD patients into UC and CD, an almost similar pattern was noticed in both the groups. The results of TNF-β (+252A/G) polymorphisms showed a significant increase in the frequency of the GG genotype in IBD patients, suggesting a positive association of GG genotype with IBD risk. On stratification of IBD patients into UC and CD, the genotype GG of TNF-β was associated with susceptibility risk to UC but not CD. The frequencies of alleles and genotypes of both TNF-α and-β polymorphisms are not affected by sex or type of IBD (familial or sporadic). TNF-α (–308G/A) and TNF-β (+252A/G) polymorphisms are associated with risk of developing IBD in Saudi population.
Keywords: tumor necrosis factor, polymorphism, inflammatory bowel disease, Saudis, Crohn’s disease, ulcerative colitis
Background
Inflammatory bowel diseases (IBDs) refer to two chronic inflammatory disorders of the gastrointestinal tract: ulcerative colitis (UC) and Crohn’s disease (CD). Increasing incidence and prevalence of IBD worldwide have made it a global disease of medical importance.1 Patients with long-lasting IBD, both UC and CD, have been at increased risk of developing colorectal cancer, and CD patients are at increased risk of small intestine cancer.2
IBD is a complex multifactorial disease in which immune dysregulation caused by genetic and/or environmental factors plays an important role.3–7 It is believed to be caused by immunogenic responses against environmental factors and/or microbes inhabiting the distal ileum and colon of genetically susceptible hosts. Various epidemiological and population-based studies have indicated that genetic factors contribute to the pathogenesis of IBD.8–12
An active inflammatory response is an important feature of IBD. Tumor necrosis factor-alpha (TNF-α) is a key cytokine in the initiation and propagation of IBD. High serum levels of TNF-α together with the increased expression of TNF-α have been documented in intestinal tissues and peripheral phagocytes of patients with IBD.13–15 It has also been shown that monoclonal antibodies against TNF-α are effective for decreasing inflammation in IBD.16 The genetic variation in TNF-a gene at position –308 results in two allelic forms in which the presence of guanine (G) defines the common variant and the presence of adenine (A) defines the less common one. TNF-α (–308) A allele displays increased gene transcription as compared to the common allele G. It has been shown to produce 6- to 7-fold higher levels of TNF-α transcription.17,18 A polymorphism at position +252 within the first intron of the TNF-b gene, consists of a guanine (TNF-β +252 G) on one allele and an adenine (TNF-β+252A) on the alternate allele. The presence of G at this position defines the mutant allele known as TNF-β*1 (allele-1), which is less frequent allele and is associated with higher TNF-α and TNF-β production.19,20 Alterations in TNF expression related to polymorphic alleles of the TNF genes have been implicated in the pathogenic role of this cytokine in several chronic inflammatory and autoimmune diseases including IBD21 and have therefore been considered an important target for interfering with the inflammatory responses.
Genetic associations have been reported between promoter polymorphisms of TNF-a and IBD.22–24 However, these associations are inconsistent and not universally replicated.25 These differences have been attributed to genetic variations of the different populations or systemic differences in the ancestry of cases and controls.15 Intriguingly, the characteristics of Western and Asian IBD patients differ in epidemiology, phenotype, and genetic susceptibility.8,26 Evaluating these unique features in different ethnic populations may provide substantial clues for identifying the pathophysiology and understanding the etiology of IBD. Saudi population being a closed and isolated society with high rate of consanguinity (inbreeding) is ideal for such genetic association studies. In this study, we determined the association of variants of TNF-α (–308G/A) and TNF-β (+252A/G) polymorphisms with the susceptibility risk to IBD in Saudi population.
Methods
Subjects
A total of 379 Saudi subjects including 179 IBD patients visiting Gastroenterology Clinic and 200 age- and sex-matched healthy controls visiting Community Health Clinic of Prince Sultan Military Medical City, Riyadh, were recruited for this study. Blood samples were obtained from all the subjects. Of the patients with IBD, 20 had the familial form and 159 had sporadic form of the disease. These were classified into 95 patients with CD (57 men and 38 women), with a mean age of 32 years (range, 17–65 years), and 84 patients with UC (34 men and 50 women), with a mean age of 34 years (range, 22–68 years). A total of 200 healthy individuals (120 men and 80 women) matched for age (mean age 30 years [range, 20–65 years]) and ethnicity (Saudis) were included as controls. None of the healthy controls had any evidence of IBD, diabetes, rheumatoid arthritis, systemic lupus erythematosus, psoriasis, or other autoimmune/inflammatory diseases. All subjects were unrelated Saudis. The diagnoses of CD and UC were determined according to conventional endoscopic, radiological, and histological criteria.27 Data obtained from each patient included age at diagnosis, disease location, disease characteristics, and extraintestinal location, which were used to group the patients. Patients with clinical features of both CD and UC (and therefore classified as “indeterminate colitis”) were excluded from this study. IBD patients with any other autoimmune/inflammatory disease were also excluded from the study. Written informed consent was obtained from all patients and controls to participate in this study, which was approved by the Ethical Committee of Prince Sultan Military Medical City, Riyadh.
Polymerase chain reaction amplification
Genomic DNA were extracted from the blood of IBD patients and controls using a QIA amp® DNA Mini Kit (Qiagen, Valencia, CA, USA). TNF-a and TNF-b genes were amplified using an amplification refractory mutation systems polymerase chain reaction (PCR) methodology (described elsewhere28) to detect polymorphisms at position –308 and intron 1 +252 of the TNF-a and TNF-b genes, respectively. PCR amplification was carried out in PuReTaq Ready-to-Go PCR Beads (GE Healthcare UK Ltd, Little Chalfont, UK). The reaction conditions consisted of ten temperature cycles of denaturation for 15 seconds at 94°C, annealing for 50 seconds at 65°C, and extension for 40 seconds at 72°C. Then, 25 cycles of denaturation for 20 seconds at 94°C, annealing for 50 seconds at 59°C, and extension for 50 seconds at 72°C were performed. A final extension was performed at 72°C for 7 minutes. A positive control was included in the PCR assay via amplification of the human growth hormone gene. For quality control, 25% of the random blind samples were repeated for genotyping and a negative control was also used in the PCR. The frequencies of alleles and genotypes were calculated in patient and control groups. Hardy–Weinberg equilibrium was determined using Hardy–Weinberg equilibrium calculator for two alleles (http//www.had2know.com/academics/hardy-weinberg-equilibrium calculator-alleles.html).
Statistical analysis
Frequencies of various alleles and genotypes for each polymorphism were compared between patients and controls and analyzed by Fisher’s exact test using the CalcFisher software (http://www.jstatsoft.org/v08/i21/paper), and P-values≤0.05 were considered as significant. The strength of the association of disease with respect to a particular allele/genotype is expressed by odds ratio interpreted as relative risk (RR) according to the method of Woolf as outlined by Schallreuter et al.29 The RR was calculated only for those alleles and genotypes that were increased or decreased in IBD patients as compared to healthy controls. RR was calculated using the following formula:
|
|
where a is the number of patients expressing the allele or genotype; b the number of patients without allele or genotype expression; c the number of controls expressing the allele or genotype; and d the number of controls without allele or genotype expression.
The etiologic fraction (EF) indicates the hypothetical genetic component of the disease. EF values of >0.00–0.99 are significant. It is calculated for positive associations (RR >1) using the following formula proposed by Svejgaard et al:30
|
|
Preventive fraction (PF) indicates the hypothetical protective effect of one allele/genotype for a disease. It is calculated for negative associations (RR <1) using the following formula:30
|
|
Values of <1.0 indicate the protective effect of an allele/genotype against the manifestation of disease.
Results
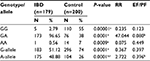
The demographic and characteristic features of IBD (CD and UC) patients are summarized in Table 1. The pathological assessment of IBD was based on two types of lesions, combined with each other in various ways, represented by architectural abnormalities (that include crypt branching/shortening, decreased crypt density, and irregular mucosal surface) and inflammatory features (transmucosal increase of lamina propria mononuclear cells and the presence of epithelioid granulomas). The extent of disease for UC and CD patients and its frequency is listed in Tables 2 and 3.
  | Table 1 Demographic and characteristic features of IBD patients Abbreviations: UC, ulcerative colitis; CD, Crohn’s disease; IBD, inflammatory bowel disease. |
  | Table 2 Extent of disease for UC patients (n=84) Abbreviations: n, number of patients; UC, ulcerative colitis. |
  | Table 3 Extent of disease for CD patients (n=95) Abbreviations: n, number of patients; CD, Crohn’s disease. |
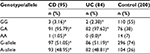
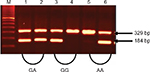
The results of the genotypes and alleles distribution of TNF-α (–308G/A) and TNF-β (+252A/G) polymorphisms in IBD and controls are summarized in Tables 4–9. The representative gel pictures for amplification of TNF-a and TNF-b are shown in Figures 1 and 2. The frequencies of alleles and genotypes of both TNF-α (–308G/A) and TNF-β (+252A/G) polymorphisms differ between IBD patients and control subjects. The allele frequencies of both patients and controls were in Hardy–Weinberg equilibrium. The frequency of GA (–308) genotype was significantly higher, while the frequencies of GG (–308) and AA (–308) genotypes were lower in IBD patients as compared to controls. Allele A (TNF-α 2-allele) of TNF-α (–308G/A) polymorphism was found to be significantly more in IBD patients, while allele G (TNF-α 1-allele) was found to be more in the control group (Table 4). Allele A and genotype GA were associated with susceptibility risk to the IBD (P<0.001, 95% confidence interval =1.66–4.45), while allele G and genotype GG might be protective against IBD (P<0.001) in Saudi patients.
On stratification of our results for IBD patients into UC and CD, an almost similar pattern was noticed in both the groups, and the difference in distribution of GG and GA genotypes of TNF-α (–308G/A) polymorphism was significant for both UC and CD as compared to controls (Table 5). Allele A and genotype GA were associated with susceptibility to the UC and CD (Ps<0.01), while allele G and genotype GG were protective (Ps<0.01) in Saudi patients with UC and CD.
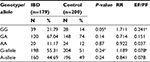
On the other hand, studies on TNF-b gene polymorphism showed that the frequency of GG at position +252 of intron 1 was significantly higher in IBD as compared to controls, while the difference in the frequencies of GA and AA genotype between patients and controls was not statistically significant (Table 6). The difference in the distribution of allele-A and allele G was also not statistically significant in IBD and control groups (P=0.24, 95% confidence interval =0.631–1.11).
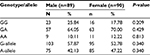
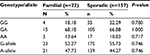
On stratification of TNF-β gene polymorphism results for UC and CD; significant difference was noticed in the distribution of genotypes GG and GA in UC, while no difference was observed in CD when compared with controls. These results indicated that the genotype GG was associated with susceptibility only for UC but not for CD as an almost similar distribution of genotypes and allele frequencies of TNF-β -intron1 +252 polymorphism was found among the CD and controls (Table 7). The frequency distribution of alleles and genotypes of both TNF-α and-β polymorphisms is not affected by sex or type of IBD (familial or sporadic; Tables 8 and 9). However, results should be interpreted with caution as the number of patients with familial form of IBD is very small (20) as compared to those with the sporadic form (159) of the disease.
Discussion
TNF-α is a key cytokine in the inflammatory response of IBD and appears to be important in the digestive and systemic manifestations of the disease. Research on the polymorphism at position –308 in the promoter region of the TNF-a gene has demonstrated its importance with regard to the clinical presentation of CD and UC.23–25 Our results indicated that GA genotype and allele A of TNF-α (–308G/A) polymorphism is associated with susceptibility risk to IBD. Similar results have been reported earlier from various other populations. The polymorphism of TNF-α (–308 G/A) modifies the susceptibility to UC and CD in Europeans and Asians. Earlier, allele A of TNF-α (–308G/A) was demonstrated to be associated with susceptibility to UC in Japanese and Han Chinese patients.22,31,32 The AA genotype increases the risk of UC and CD significantly in European patients, while the GA genotype increases the risk of UC in Asians.23 Another systemic review and meta-analysis also indicated an association of TNF-α (308G/A) polymorphism with IBD in Asians and suggested that genetic mutations of IBD in Asians differ from Caucasians.24
Various reports suggested a significant association of TNF-α (–308G/A) polymorphism with IBD severity. TNF-α (–308G/A) polymorphism is significantly involved in the severity to CD and/or UC in Irish,33 Czech,34 Italian,35 and Caucasian patients from New Zealand36 and in Brazilian patients.25 It has also been suggested that the TNF-α (–308G/A) polymorphism may either be directly involved in pathophysiology of IBD or serve merely as markers in linkage disequilibrium with susceptibility genes in Irish patients.33 Individuals carrying the allele-A of TNF-α (–308G/A) polymorphism have a significantly greater risk of pancolitis and are also more likely to require bowel resection in UC and CD.34,35 Patients with CD carrying A allele were more frequently resistant to steroids compared with noncarriers, and both UC and CD patients carrying the A allele show a significant increase in CRP level, which plays a role in modifying the IBD phenotype, influencing disease activity, leading to a more intense inflammatory activity.35
The A-containing genotypes of TNF-α (–308G/A) have also been associated with degree of inflammation, with increased levels of C-reactive protein, TNF-α, and interleukin 1β in the active phase of IBD.37–40 Kim et al41 also reported higher frequency of –308A allele of TNF-α in antineutrophil cytoplasmic antibodies-positive IBD patients than antineutrophil cytoplasmic antibodies-negative IBD patients and suggested that the TNF-α (–308G/A) polymorphisms may have influences on the susceptibility to CD or the behavior of CD. On the other hand, Ferreira et al42 reported that AA genotype of TNF-α (–308) is associated with susceptibility to CD in Portuguese patients and suggested that TNF-α (–308G/A) polymorphism is responsible for displaying distinct clinicopathological profiles.
TNF-α (–308) A allele increases gene transcription as compared to the common allele G and produces a six- to sevenfold higher level of TNF-α transcription.17,18 Elevated TNF-α may play a pivotal role in the pathogenesis of the inflammatory response by interacting with TNF receptor and cause apoptosis, cell proliferation, and differentiation.43 Moreover, TNF-α overproduction has been implicated in a variety of symptoms associated with autoimmune disorders, including IBD, and especially CD.21 Inflammation, anorexia, and weight loss are also all associated with increased levels of circulating TNF-α.44
In contrast, some reports indicated lower frequency of A allele of TNF-α (–308G/A) in CD or UC in North European Caucasian and Korean patients as compared to healthy controls.45,46 Recently, Bonyadi et al47 reported that the frequency of allele G was slightly higher in Iranian Azari Turkish IBD patients, but this did not reach statistical significance. Furthermore, there are reports indicating a lack of association of TNF-α (–308G/A) polymorphism with susceptibility to IBD in Australian,48 Brazilian,25,49 Canadian,50 Chinese,32,51 Czech,52 French,53 Indian,54 Korean,5 Newfoundland,15 Spanish,55 and Turkish56 populations. These differences in associations of TNF-α (–308G/A) polymorphism with IBD might be due to variations in sample size, genotyping methods, and/or ethnicity, as frequencies of TNF (–308) alleles and genotypes also vary in different ethnic healthy populations worldwide.28
Our results also indicated that GG genotype at position +252 of intron 1 of TNF-β was significantly associated with IBD, while the frequency of GA genotype was slightly lower in IBD patients than the controls, but this difference did not reach statistical significance. However, upon stratification of the patients into UC and CD, it was noticed that the TNF-β (+252A/G) polymorphism was significantly associated with UC but not with the CD in the Saudi population (Table 7). Contrary to our findings, no correlation was reported between TNF-β (+252) polymorphism and CD or UC in Chinese, French, Korean, and Spanish patients.5,22,32,53,57 It is possible that the TNF-β (+252A/G) polymorphism maybe indirectly associated with IBD as it has been suggested that allele G influences the expression/production of TNF-α.19 Muro et al58 suggested that TNF-α and -β play an important role in inflammatory response, and IBD is commonly treated with TNF-α inhibitors. Additionally, as polymorphisms of TNF-α gene affect the gene expression level, particular TNF-α genotypes may influence the response of IBD patients treated with TNF-α inhibitors. The promoter allele A of TNF-α (–308G/A) and allele G of TNF-β (+252A/G) have been shown to be associated with greater TNF-α transcription.17,19,59,60
This study suggested a significant association between allele frequency and genotype distribution of TNF-α (–308G/A) and TNF-β (+252A/G) polymorphisms and IBD susceptibility risk in Saudi population. However, there was no correlation between TNF-α and TNF-β polymorphisms and sex or type of IBD in our population. These results are in accordance with the earlier reports from Chinese population, indicating that the polymorphisms of the TNF-α –308 and TNF-β+252 do not correlate with age, sex, disease activity, or lesion site.22,32 This is the first report from a Saudi population showing the association of TNF-α and TNF-β polymorphisms in the etiology of UC and CD. These results together with other published reports supported an important role of ethnicity in the association of TNF-α and TNF-β polymorphism and IBD. It can also be concluded that TNF-α (–308G/A) and TNF-β (+252A/G) polymorphisms may work together in the pathogenesis of IBD in Saudi population. The results of this study may have prognostic value for future clinical observations of Saudi IBD patients, and TNF-α (–308G/A) polymorphism may help in determining the response to anti-TNF-α therapy as patients with different genotype respond differently to anti-TNF-α treatment.61,62 However, further studies are required involving other ethnic populations to confirm association of these polymorphisms with susceptibility of IBD.
Acknowledgments
The authors thank S Sadaf Rizvi and Mohammad Al-Asmari for their help with laboratory work.
Disclosure
The authors report no conflicts of interest in this work.
References
Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. | ||
Jussila A, Virta LJ, Pukkala E, Färkkilä MA. Mortality and causes of death in patients with inflammatory bowel disease: a nationwide register study in Finland. J Crohns Colitis. 2014;8:1088–1096. | ||
Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. | ||
Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. | ||
Yang SK, Lee SG, Cho YK, Lim J, Lee I, Song K. Association of TNF-a/LTA polymorphisms with Crohn’s disease in Koreans. Cytokine. 2006;35:13–20. | ||
Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–552. | ||
Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. | ||
Kim ES, Kim WH. Inflammatory bowel disease in Korea: epidemiological, genomic, clinical, and therapeutic characteristics. Gut Liver. 2010;4:1–14. | ||
Yun J, Xu CT, Pan BR. Epidemiology and gene markers of ulcerative colitis in the Chinese. World J Gastroenterol. 2009;15:788–803. | ||
Lowe AM, Roy PO, B-Poulin M, et al. Epidemiology of Crohn’s disease in Québec, Canada. Inflamm Bowel Dis. 2009;15:429–435. | ||
Agouridis AP, Elisaf M, Milionis HJ. An overview of lipid abnormalities in patients with inflammatory bowel disease. Ann Gastroenterol. 2011;24:181–187. | ||
Waterman M, Xu W, Stempak JM, et al. Distinct and overlapping genetic loci in Crohn’s disease and ulcerative colitis: correlations with pathogenesis. Inflamm Bowel Dis. 2011;17:1936–1942. | ||
Plevy SE, Landers CJ, Prehn J, et al. A role for TNF-alpha and mucosal T helper-1 cytokines in the pathogenesis of Crohn’s disease. J Immunol. 1997;159:6276–6282. | ||
Komatsu M, Kobayashi D, Saito K, et al. Tumor necrosis factor-alpha in serum of patients with inflammatory bowel disease as measured by a highly sensitive immuno-PCR. Clin Chem. 2001;47:1297–1301. | ||
Zipperlen K, Peddle L, Melay B, Hefferton D, Rahman P. Association of TNF-alpha polymorphisms in Crohn disease. Hum Immunol. 2005;66:56–59. | ||
Deleporte A, Viennot S, Dupont B, et al. Efficacy of anti-TNF-alpha monoclonal antibodies in inflammatory bowel disease treatment. Int J Interferon Cytokine Mediator Res. 2013;5:11–31. | ||
Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A. 1997;94:3195–3099. | ||
Abdallah AN, Cucchi-Mouillot P, Biteau N, Cassaigne A, Haras D, Iron A. Analysis of the polymorphism of the tumour necrosis factor (TNF) gene and promoter and of circulating TNF-alpha levels in heart-transplant patients suffering or not suffering from severe rejection. Eur J Immunogenet. 1999;26:249–255. | ||
Messer G, Spengler U, Jung MC, et al. Polymorphic structure of the tumor necrosis factor (TNF) locus: an Ncol Polymorphism in the first intron of the human TNF-beta gene correlates with a variant amino acid in position 26 and a reduced level of TNF-beta production. J Exp Med. 1991;173:209–219. | ||
Abraham LJ, Kroeger KM. Impact of the -308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol. 1999;66:562–566. | ||
Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214: | ||
Cao Q, Zhu Q, Wu ML, Hu WL, Gao M, Si JM. Genetic susceptibility to ulcerative colitis in the Chinese Han ethnic population: association with TNF polymorphisms. Chin Med J (Engl). 2006;119:1198–1203. | ||
Fan W, Maoqing W, Wangyang C, et al. Relationship between the polymorphism of tumor necrosis factor-α-308 G>A and susceptibility to inflammatory bowel diseases and colorectal cancer: a meta-analysis. Eur J Hum Genet. 2011;19:432–437. | ||
Ng SC, Tsoi KK, Kamm MA, et al. Genetics of inflammatory bowel disease in Asia: systematic review and meta-analysis. Inflamm Bowel Dis. 2012;18:1164–1176. | ||
Santana G, Bendicho MT, Santana TC, Reis LB, Lemaire D, Lyra AC. The TNF-α -308 polymorphism may affect the severity of Crohn’s disease. Clinics (Sao Paulo). 2011;66:1373–1377. | ||
Leong RW, Lau JY, Sung JJ. The epidemiology and phenotype of Crohn’s disease in the Chinese population. Inflamm Bowel Dis. 2004;10:646–651. | ||
Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2–6; discussion 16–19. | ||
Al-Rayes H, Al-Swailem R, Albelawi M, Arfin M, Al-Asmari A, Tariq M. TNF-α and TNF-β gene polymorphism in Saudi rheumatoid arthritis patients. Clin Med Insights Arthritis Musculoskelet Disord. 2011;4:55–63. | ||
Schallreuter KU, Levenig C, Kühnl P, Löliger C, Hohl-Tehari M, Berger J. | ||
Svejgaard A, Platz P, Ryder LP. HLA and disease 1982 – a survey. Immunol Rev. 1983;70:193–218. | ||
Sashio H, Tamura K, Ito R, et al. Polymorphisms of the TNF gene and the TNF receptor super family member 1B gene are associated with susceptibility to ulcerative colitis and Crohn’s disease, respectively. Immunogenetics. 2002;53:1020–1027. | ||
Song Y, Wu KC, Zhang L, et al. Correlation between a gene polymorphism of tumor necrosis factor and inflammatory bowel disease. Chin J Dig Dis. 2005;6:170–174. | ||
Balding J, Livingstone WJ, Conroy J, et al. Inflammatory bowel disease: the role of inflammatory cytokine gene polymorphisms. Mediators Inflamm. 2004;13:181–187. | ||
Sykora J, Subrt I, Didek P, et al. Cytokine tumor necrosis factor-alpha A promoter gene polymorphism at position -308 G-->A and pediatric inflammatory bowel disease: implications in ulcerative colitis and Crohn’s disease. J Pediatr Gastroenterol Nutr. 2006;42: | ||
Cucchiara S, Latiano A, Palmieri O, et al. Polymorphisms of tumor necrosis factor alpha but not MDR1 influence response to medical therapy in pediatric-onset inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2007;44:171–179. | ||
Ferguson LR, Huebner C, Petermann I, et al. Single nucleotide polymorphism in the tumor necrosis factor-alpha gene affects inflammatory bowel diseases risk. World J Gastroenterol. 2008;14:4652–4661. | ||
Louis E, Peeters M, Franchimont D, et al. Tumor necrosis factor (TNF) gene polymorphism in Crohn’s disease (CD): influence on disease behavior? Clin Exp Immunol. 2000;119:64–68. | ||
Louis E, Vermeire S, Rutgeerts P, et al. A positive response to infliximab in Crohn disease: association with a higher systemic inflammation before treatment but not with -308 TNF gene polymorphism. Scand J Gastroenterol. 2002;37:818–824. | ||
Vatay A, Bene L, Kovacs A, et al. Relationship between the tumor necrosis factor alpha polymorphism and the serum C-reactive protein levels in inflammatory bowel disease. Immunogenetics. 2003;55: | ||
Gonzalez S, Rodrigo L, Martinez-Borra J, et al. TNF-alpha -308A promoter polymorphism is associated with enhanced TNF-alpha production and inflammatory activity in Crohn’s patients with fistulizing disease. Am J Gastroenterol. 2003;98:1101–1106. | ||
Kim TH, Kim BG, Shin HD, et al. Tumor necrosis factor-alpha and interleukin-10 gene polymorphisms in Korean patients with inflammatory bowel disease. Korean J Gastroenterol. 2003;42:377–386. | ||
Ferreira AC, Almeida S, Tavares M, et al. NOD2/CARD15 and TNFA, but not IL1B and IL1RN, are associated with Crohn’s disease. Inflamm Bowel Dis. 2005;11:331–339. | ||
Stokkers PC, Hommes DW. New cytokine therapeutics for inflammatory bowel disease. Cytokine. 2004;28(4–5):167–173. | ||
Muro M, López-Hernandez R, Campillo JA, et al. Recent advances in basic and clinical aspects in inflammatory bowel diseases, as Crohn’s disease and ulcerative colitis: role of gastrointestinal granulocytes. In: Berhardt LV, editor. Advances in Medicine and Biology. New York, NY: Nova Science Publishers, Inc.; 2010:133–154. | ||
Bouma G, Xia B, Crusius JB, et al. Distribution of four polymorphisms in the tumor necrosis factor (TNF) genes in patients with inflammatory bowel disease (IBD). Clin Exp Immunol. 1996;103:391–396. | ||
Louis E, Satsangi J, Roussomoustakaki M, et al. Cytokine gene polymorphism in inflammatory bowel disease. Gut. 1996;39:705–710. | ||
Bonyadi M, Abdolmohammadi R, Jahanafrooz Z, Somy MH, Khoshbaten M. TNF-alpha gene polymorphisms in Iranian Azari Turkish patients with inflammatory bowel diseases. Saudi J Gastroenterol. 2014;20:108–112. | ||
Fowler EV, Eri R, Hume G, et al. TNF alpha and IL10 SNPs act together to predict disease behaviour in Crohn’s disease. J Med Genet. 2005; | ||
Queiroz DM, Oliveira AG, Saraiva IE, et al. Immune response and gene polymorphism profiles in Crohn’s disease and ulcerative colitis. Inflamm Bowel Dis. 2009;15:353–358. | ||
Cantor MJ, Nickerson P, Bernstein CN. The role of cytokine gene polymorphisms in determining disease susceptibility and phenotype in inflammatory bowel disease. Am J Gastroenterol. 2005;100:1134–1142. | ||
Han Z, Li C, Han S, et al. Meta-analysis: polymorphisms in TNF-alpha gene promoter and Crohn’s disease. Aliment Pharmacol Ther. 2010;32:159–170. | ||
Hradsky O, Lenicek M, Dusatkova P, et al. Variants of CARD15, TNFA and PTPN22 and susceptibility to Crohn’s disease in the Czech population: high frequency of the CARD15 1007fs. Tissue Antigens. 2008;71:538–547. | ||
Heresbach D, Ababou A, Bourienne A, et al. Polymorphism of the microsatellites and tumor necrosis factor genes in chronic inflammatory bowel diseases. Gastroenterol Clin Biol. 1997;21:555–561. | ||
Mittal RD, Manchanda PK, Bid HK, Ghoshal UC. Analysis of polymorphisms of tumor necrosis factor-alpha and polymorphic xenobiotic metabolizing enzymes in inflammatory bowel disease: study from northern India. J Gastroenterol Hepatol. 2007;22:920–924. | ||
Castro-Santos P, Suarez A, Lopez-Rivas L, Mozo L, Gutierrez C. TNF alpha and IL-10 gene polymorphisms in inflammatory bowel disease. Association of -1082 AA low producer IL-10 genotype with steroid dependency. Am J Gastroenterol. 2006;101:1039–1047. | ||
Celik Y, Dagli U, Kiliç MY, et al. Cytokine gene polymorphisms in Turkish patients with inflammatory bowel disease. Scand J Gastroenterol. 2006;41:559–565. | ||
Papo M, Quer JC, Gutierrez C, et al. Genetic heterogeneity within ulcerative colitis determined by an interleukin-1 receptor antagonist gene polymorphism and antineutrophil cytoplasmic antibodies. Eur J Gastroenterol Hepatol. 1999;11:413–420. | ||
Muro M, López-Hernández R, Mrowiec A. Immunogenetic biomarkers in inflammatory bowel diseases: role of the IBD3 region. World J Gastroenterol. 2014;20:15037–15048. | ||
Brinkman BM, Giphart MJ, Verhoef A, et al. Tumor necrosis factor alpha-308 gene variants in relation to major histocompatibility complex alleles and Feltys syndrome. Hum Immunol. 1994;41:259–266. | ||
Jeong P, Kim EJ, Kim EG, Byun SS, Kim CS, Kim WJ. Association of bladder tumors and GA genotype of -308 nucleotide in tumor necrosis factor-alpha promoter with greater tumor necrosis factor-alpha expression. Urology. 2004;64:1052–1056. | ||
Peyrin-Biroulet L. Anti-TNF therapy in inflammatory bowel diseases: a huge review. Minerva Gastroenterol Dietol. 2010;56:233–243. | ||
Lopez A, Billioud V, Peyrin-Biroulet C, Peyrin-Biroulet L. Adherence to anti-TNF therapy in inflammatory bowel diseases: a systematic review. Inflamm Bowel Dis. 2013;19:1528–1533. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.