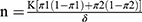Back to Journals » Journal of Pain Research » Volume 14
Association of Single Nucleotide Polymorphism rs2228570 with Lumbar Disc Degeneration: A Case–Control Study and Meta-Analysis
Authors Zhang H , Chen L, Wang Z , Wang F , Shan Y, Qi L, Chen Y
Received 1 April 2021
Accepted for publication 17 June 2021
Published 1 July 2021 Volume 2021:14 Pages 2001—2012
DOI https://doi.org/10.2147/JPR.S313790
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor E Alfonso Romero-Sandoval
Hengshuo Zhang, Lu Chen, Ziyu Wang, Fuan Wang, Yu Shan, Linzeng Qi, Yunzhen Chen
Department of Spine Surgery, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Jinan, People’s Republic of China
Correspondence: Yunzhen Chen Email [email protected]
Objective: To examine the association between single nucleotide polymorphisms (SNPs) rs2228570, rs731236, rs7975232, and rs1544410 and lumbar disc degeneration (LDD) predisposition.
Methods: A search strategy was carried out, and the data were extracted after being chosen by the inclusion and exclusion criteria. Pooled odds ratios and 95% confidence intervals were calculated to assess the association between the aforementioned SNPs and LDD under allelic, dominant, recessive, heterozygous, and homozygous genetic models. In addition, a case–control study involving 46 LDD cases and 45 controls was also performed in the analysis to verify the result.
Results: A total of 17 studies were included in this meta-analysis. The pooled results did not show any association between vitamin D receptor (VDR) gene polymorphisms and LDD. But, interestingly, in subgroup analysis, the rs2228570 polymorphism was associated with LDD under the allelic (OR = 0.70, 95% CI = 0.56– 0.87, p = 0.002), recessive (OR = 0.60, 95% CI = 0.43– 0.84, p = 0.003), and homozygous (OR = 0.47, 95% CI= 0.28– 0.79, p = 0.004) genetic models in the Asian population. SNPs rs731236 and rs7975232 still did not show any obvious association. We obtained a similar result from the case–control study: rs2228570 had an obvious relationship with LDD under allelic and homozygous genetic models. At the same time, we found that rs2228570 was also associated with the degree of low back pain (visual analogue scale, VAS score) in LDD population.
Conclusion: SNP rs2228570 was significantly associated with LDD predisposition and the degree of low back pain in the Asian population.
Keywords: vitamin D receptor, single nucleotide polymorphism, lumbar disc degeneration, visual analogue scale, meta-analysis, case-control study
Introduction
Low back pain (LBP) is the third most common disorder in neurology, following peripheral neuropathy and migraine, and thus affects a large population. The pain might arise from nerves, muscles,1 or bones in the back, and various etiological factors may contribute to the pain. Among these factors, lumbar disc degeneration (LDD) is considered a significant cause.2
In the pathogenesis of LDD, both environmental and genetic factors play important roles. Environmental factors include physical factors or psychosocial factors,3 and in terms of genetic factors, single nucleotide polymorphisms (SNPs) have been recognized to be significant recently. It seems that SNPs in the genes of disc proteins, proteoglycans, cytokines, enzymes, and vitamin D receptor (VDR) would contribute considerably to an LDD attack.4 Among them, VDR gene polymorphisms have been widely explored to assess the association with LDD.
VDR is a nucleoprotein, and it is crucial for maintaining the body’s calcium and phosphorus metabolism and regulating cell proliferation and differentiation. As a candidate gene for LDD, VDR gene, which is located on human chromosome 12 with a length of 100 kb, has been widely studied by many scientific research groups. Up till now, the association of several VDR gene polymorphisms, including rs2228570, rs731236, rs7975232, and rs1544410, with LDD predisposition has been reported.5 However, the results are contradictory. Several studies have suggested that there are obvious associations between VDR gene polymorphisms and the risk of LDD,6–8 whereas some other studies have reported contrary conclusions.9–11 With inconsistent results from these studies based on relatively small sample sizes, we cannot draw a valid conclusion. Although several systematic reviews and meta-analysis have summarized, integrated, and analyzed previous studies, considering that there are several new studies and some deviations in collection of data from previous studies, it is necessary to update the meta-analysis and obtain a more reliable result with a more accurate and comprehensive analysis. In this study, we conducted a systematic review and meta-analysis of VDR gene polymorphisms and risk of LDD. We used normal people’s and patients’ genetic information to verify the meta-analysis result.
Materials and Methods
Study Subjects
From March 2019 to May 2019, 46 patients with confirmed LDD and 45 normal controls were recruited from the Qilu Hospital of Shandong University. A total of 2 mL peripheral venous blood from the selected population was collected and used to isolate and extract genome-wide DNA from individuals for further detection. Basic information like age, gender, height, weight and special information in case group like VAS score and imaging data were also collected. This study was approved by the Ethics Committee of Qilu Hospital of Shandong University. Inclusion criteria were as follows: (1) The age of the participants should be greater than 18 years. (2) The patients in the case group were definitely diagnosed with lumbar disc degeneration, with single central disc herniation and required surgical treatment. (3) The selected candidate could undergo surgery under general anesthesia. (4) Candidates should have normal mental consciousness and be able to cooperate with relevant research. (5) Candidates should be willing to participate in the study and sign the informed consent. Exclusion criteria were as follows: (1) Pregnant and lactating patients, infants, and adolescents; (2) Cannot cooperate with researchers; (3) A history of spinal trauma; (4) Patients with scoliosis, spinal deformity, metabolic diseases, and malignant tumors; (5) Have undergone surgery in the same period.
Sample Size Calculation
where12
n = the sample size in one group and K = a function of the chosen significance level and chosen power.
The power of the study will be 80% and the two-sided significance level will be 0.05. The K value is for the power and the significance level is 7.8.
The characteristic of interest—polymorphism of the VDR gene.
π1 = Proportion of subjects with the disease who are having the characteristic of interest.
π2 = Proportion of normal subjects who are having the characteristic of interest.
δ = (π1-π2).
n= 45 (each in test and control).
DNA Extraction and Genotyping
Blood samples were digested by SDS/proteinase K (QIAGEN, Duesseldorf, Germany), the activity of the proteinase K solution is 600 mAU/mL. The DNA of samples was extracted by phenol/chloroform, and then precipitated with ethanol. Finally, the DNA was dissolved in a Reduced EDTA TE Buffer at a concentration of no less than 50 ng/μL and a total of no less than 1 μg. Agarose gel electrophoresis was performed on the sample DNA, and the result showed clear electrophoresis bands with a length more than 10 kb and there was no significant DNA degradation. OD 260/280 of DNA sample was between 1.7 and 2.1. In this study, genotyping was performed by using Capital Biotechnology Precision Medicine Research Array Kit (Thermo Fisher Scientific, Waltham, MA, USA), which is a customized chip based on Axiom 2.0 platform. This microarray contains over 787,400 SNPs, which includes 50,000 novel markers covering East and South Asian populations based on the human genome version 19 (GRCH 37).
Low Back Pain VAS Score Assessment
Low back pain degree of 46 patients with confirmed LDD was assessed before blood collection by using the Visual Analogue Scale (VAS): 0 points, no pain; 1–3 points, slight pain, tolerable; 4–6 points, moderate pain, interfere with sleep, tolerable; 7–10 points, severe pain, interfere with sleep and appetite, unbearable.
Statistical Analysis
For the case–control study, the χ2 test was performed to assess the Hardy–Weinberg equilibrium (HWE) in control cases. For each analysis, the results were considered to be statistically significant when two-sided P was <0.05.
Meta-Analysis
Search Strategy
PubMed, EMBASE, COCHRANE, and three Chinese databases—CNKI, Wanfang, and VIP—were searched up to May 31, 2019. The reference lists of some related articles were screened, as well as those of conference articles and clinical trials. The detailed search strategy is shown in the supplement.
Study Selection and Data Extraction
This work was independently completed by three authors (Lu Chen, Ziyu Wang and Fuan Wang). Any disagreement was settled by consultation. Data obtained included the first author, year of publication, country where the study was conducted, number of participants in the case and control groups, number of male and female participants, age and BMI of the participants, and the method for diagnosis and genotyping. The criteria for inclusion were as follows: (1) Case–control study or cohort study, (2) Patients selected on the basis of specific diagnostic criteria, (3) Complete data. The criteria for exclusion were as follows: (1) Duplication, (2) Defect of the study design, (3) No specific diagnostic criteria, (4) Errors in the statistical method or no supplement for the data that was or could be converted to OR and 95% CI.
Statistical Analysis
Meta-analysis was performed by Review Manager 5.3 (the Cochrane Collaboration, UK) and STATA 12.0 (StataCorp LLC, US). The HWE test was performed in the control group of each included study, and p value >0.05 was regarded as fitting the genetic equilibrium. Summary Odds Ratios (ORs) with 95% confidence intervals (CIs), which were calculated using a fixed-effect model or a random-effect model according to the heterogeneity, were estimated for the association between the risk of LDD and SNP. Heterogeneity was reflected in I2 statistics, which is on the basis of Q test and has higher sensitivity. If I2 > 50%, the meta-analysis has obvious heterogeneity, whereas I2 < 50% means lower heterogeneity that can be accepted. Sensitivity analysis was performed using STATA 12.0, and the results showed a change in summary ORs after removing each study. Finally, the funnel plots, Begg’s test, and Egger’s test were used to measure the publication bias. The funnel plots were recommended to be performed only if more than 10 studies were included. For the case-control study, the Chi-square test was used to assess the relationship between two groups.
Results
Search Results and Characteristics of Included Studies
The literature search yielded 149 records (Figure 1), and 17 records met the inclusion criteria. Among these included studies, sample size ranged from 60 to 538 (age range, 18 to 86 years). All the studies were case–control studies and aimed at LDD. Patients were diagnosed by computed tomography (CT), magnetic resonance imaging (MRI) or both, and polymerase chain reaction (PCR) or PCR-restriction fragment length polymorphism (PCR-RFLP) was a common method for genotyping. In terms of ethnicity, eight studies were based on the Asian population, and the other studies were based on Caucasian population. Ten studies of rs2228570, nine studies of rs731236, and six studies of rs7975232 were included in the meta-analysis, and the characteristics of each study are listed in Table 1. All the records had good quality after assessment by the Newcastle−Ottawa Scale (Table S1 in the Supplement). The number of participants in the case and control groups and the types of polymorphisms in each study are listed in Table 2.
 |
Table 1 Characteristics of Studies Included in Meta-Analysis |
 |
Table 2 Distribution of Genotype and Allele of Different VDR Gene Polymorphism |
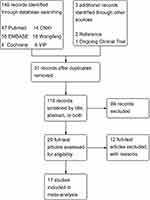 |
Figure 1 Flow diagram of studies selection. |
Quantitative Synthesis and Sensitivity Analysis
As shown in Table 3, we performed the data synthesis of the above three polymorphisms according to the following five genetic models: allelic model, dominant model, recessive model, heterozygous model, and homozygous model. All the pooled results suggested that the three polymorphisms rs2228570, rs731236, and rs7975232 had no association with LDD, but they had obvious heterogeneity except the rs2228570 heterozygous model (I2 = 34%) and the rs731236 homozygous model (I2= 0%). It is worth mentioning that there were four studies related to rs731236 that had two groups with no person under dominant, heterozygous, and homozygous genetic models; therefore, they could not be included in the pooled OR calculation. Next, we performed sensitivity analysis (Figure 2), and the results showed that the removal of either one could affect the pooled ORs significantly, which proved the high stability of the meta-analysis. That was the reason why we included the studies that did not fit the HWE.
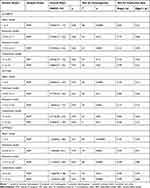 |
Table 3 Meta-Analysis of VDR Gene Polymorphisms Under Different Genetic Model |
Subgroup Analysis
To further explore the cause of heterogeneity, subgroup analysis according to the ethnicity was performed after sensitivity analysis. The data were stratified into two ethnic groups: Asian and Caucasian. For rs2228570, the heterogeneity in the Asian population decreased significantly while that in the Caucasian population showed minimal change, as well as in the rs7975232 subgroup analysis. Regarding rs731236, a contrary result was obtained; the heterogeneity in the Caucasian population decreased drastically, but it did not show any obvious association with LDD in either subgroup. It should be noted that subgroup analysis showed that rs2228570 polymorphism was associated with LDD under the allelic (Figure 2A), recessive (Figure 2B), and homozygous (Figure 2C) genetic models in the Asian population.
Publication Bias
With respect to rs2228570, 10 studies were included in this meta-analysis; therefore, funnel plots, Begg’s test, and Egger’s test were used to assess the publication bias. With respect to rs731236 and rs7975232, the number of studies was less than 10; therefore, the Begg’s test and Egger’s test were only used. All the results showed that there was no obvious publication bias in the meta-analysis of three VDR gene polymorphisms under every genetic model.
Basic Information of Research Subjects in Case–Control Study
A total of 46 patients were included in the case group, and 45 patients were included in the control group. There were no significant differences in gender, age, height, weight, and BMI between the two groups (Table S2 in the Supplement). According to the results of HWB test, P values of SNPs rs2228570, rs731236, rs7975232, and rs1544410 of the VDR gene were greater than 0.05, suggesting that their distribution in the study population met the Hardy−Weinberg balance (Table S3 in the Supplement), and the samples were representative of the population.
Distribution of Single Nucleotide Polymorphisms of VDRs and Their Relationship with Degeneration of the Lumbar Intervertebral Disc
Single Nucleotide Polymorphism rs2228570
The genotype frequency in the control group was (1 for the main allele and 2 for the suballele) 11:13.3%, 12:48.9%, and 22:37.8%. The genotype frequency in the case group was 11:28.3%, 12:52.2%, and 22:19.6%. The results showed that in the allele model (OR = 0.51, P= 0.02497) and the homozygous model (OR = 0.63, P = 0.02504), SNP rs2228570 was significantly associated with the risk of LDD (Table 4).
 |
Table 4 The Relationship Between VDR Gene Polymorphisms and Intervertebral Disc Degeneration |
Single Nucleotide Polymorphism rs731236
The genotype frequency in the control group was 11:86.7%, 12:13.3%, and 22:0%. The genotype frequency in the case group was 11:9 5.7%, 12:4.3%, and 22:0%. Under the five gene models, the distribution of rs731236 and the χ2 test value of LDD were greater than 0.05, indicating that there was no significant correlation between rs731236 and LDD (Table 4).
Single Nucleotide Polymorphism rs7975232
The genotype frequency in the control group was 11:68.9%, 12:31.1%, and 22:0%. The genotype frequency in the case group was 11:69.6%, 12:28.3%, and 22:2.2%. Under the five gene models, the distribution of rs7975232 and the χ2 test value of LDD were greater than 0.05, indicating that there was no significant correlation between rs7975232 and LDD (Table 4).
Single Nucleotide Polymorphism rs1544410
The genotype frequency in the control group was 11:91.1%, 12:8.9%, and 22:0%. The genotype frequency in the case group was 11:9 7.8%, 12:2.2%, and 22:0%. Under the five gene models, the distribution of rs1544410 and the χ2 test value of LDD were greater than 0.05, indicating that there was no significant correlation between rs1544410 and LDD (Table 4).
Relationship Between Single Nucleotide Polymorphisms of VDRs and Low Back Pain
According to the VAS score, 46 patients were divided into two groups (VAS < 4 and VAS ≥ 4). There were no significant differences in gender, age, height, weight, BMI, spinal canal occupation ratio (SCOR) and decrease in spinal canal ratio (DSCR) between the two groups (Table S4 in the Supplement). Low back pain VAS scores are shown in Table 5. SNPs rs2228570 (P=0.033) and rs7975232 (P=0.046) of the VDR gene had an obvious relationship with low back pain VAS score. However, SNPs rs731236 (P=0.23) and rs1544410 (P=0.40) did not show any significant association.
 |
Table 5 The Relationship Between VDR Gene Polymorphisms and Back Pain VAS Score |
Discussion
Over the past few years, many researchers have paid attention to the association between SNPs and diseases. In terms of the association between SNP rs2228570, rs731236, rs7975232, and rs1544410 and LDD, some studies have shown that there is no association between them, whereas other studies have reported contradictory conclusions. Therefore, we performed this meta-analysis.
From the result of this meta-analysis, it can be seen that rs2228570 was likely associated with LDD in the Asian population, and rs731236 had no association with LDD. However, we did not obtain any specific conclusion about rs7975232 from the result, because the results under different genetic models were contradictory to each other even though the heterogeneity was decreased after subgroup analysis. The small sample size might be the main cause. Studies about rs1544410 polymorphism were also collected, but there were only two studies, and one of them did not fit the HWE (p<0.0001). Therefore, the meta-analysis about rs1544410 was abandoned.
In this work, no publication bias was found and the results were very stable after sensitivity analysis. The main problem was heterogeneity. With respect to rs2228570 and rs7975232, there was no heterogeneity in the Asian population, while little change in the heterogeneity was found in the Caucasian population after subgroup meta-analysis. Besides, subgroup analysis based on other aspects, such as continents or the number of total participants, was also performed, but the heterogeneity of the Caucasian population did not decrease either, and meta-analysis based on the ethnicity seemed to be the best method. We guess that the main reason may be that some of the characteristics of Caucasian population are affected by other races due to its high mobility. After subgroup analysis, the result in the Asian population is available as a suggestion. With respect to some previous meta-analysis on this subject, the process to explore and explain the heterogeneity cannot be found.
For determination of the genetic model, we tried to use the method introduced by Thakkinstian et al.13 But the standard is very rigorous, and the realistic model may not fit very well; therefore, we have summarized and explained all the results under the five genetic models above.
In this meta-analysis, the number of included studies was the highest and the source of heterogeneity was also explored further. The whole process was integrated, clear, and logical. It is more important that we obtained a new conclusion, the rs2228570 polymorphism was associated with LDD in the Asian population, after sorting out the included studies and checking the methods of genotype specification.
Next, we conducted a case–control study on the correlation between single nucleotide polymorphism of VDRs (rs2228570, rs731236, rs7975232, and rs1544410) and the risk of LDD in the population. After calculating the sample size, we recruited 46 patients with LDD and 45 normal control volunteers, collected their basic information and blood samples, and assessed 46 patients’ low back pain VAS scores. We determined the genotype of each participant at the four polymorphic sites described above by using a single nucleotide polymorphic chip. Under the five genetic models of the allele model, dominant model, recessive model, heterozygous model, and homozygous model, we used the χ2 test to analyze the association between the genotype of polymorphism and the risk of LDD. The results showed that under the allelic model and the dominant model, the single nucleotide polymorphism rs2228570 was significantly associated with the risk of LDD, while the other three models showed no significant association with LDD. The participants included in the case–control study were all Asians; therefore, the conclusion of the case–control study was consistent with our meta-analysis: in the Asian population, rs2228570 SNP of VDR was significantly associated with the risk of LDD. In addition, after analyzing the VAS scores of patients, we found that SNP rs2228570 was also significantly correlated with the degree of low back pain.
It is worth mentioning that intervertebral disc degeneration is not an isolated process,14–17 it’s always associated with Modic change in end plate, which is also related with VDR polymorphisms,18 and fatty infiltration in paraspinal muscles. Finally, all of this may contribute to lower back pain. This is why some SNPs of VDR are associated with intervertebral disc degeneration, like rs2228570, but some do not.
Based on the result of this meta-analysis and case–control study, we think that future clinical studies need to be designed to collect more basic information and the data of clinical indicators. It will be better to explore the correlation between different SNPs and LDD or its relevant clinical indicator in different subgroups, such as age, gender, pain scores, and segment of LDD.
Conclusions
In summary, we confirmed that SNP rs2228570 was significantly associated with the risk of LDD in the Asian population, while rs731236, rs7975232 and rs1544410 have no significant correlation with LDD. Meanwhile, SNP rs2228570 was also significantly associated with the degree of low back pain in patients with LDD. Further studies will help to improve the value of the results of this study.
Abbreviations
SNPs, single nucleotide polymorphisms; LDD, lumbar disc degeneration; VDR, vitamin D receptor; VAS, visual analogue scale; LBP, low back pain; HWE, Hardy–Weinberg equilibrium; ORs, odds ratios; CIs, confidence intervals; CT, computed tomography; MRI, magnetic resonance imaging; PCR, polymerase chain reaction; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; SCOR, spinal canal occupation ratio; DSCR, decrease in spinal canal ratio; FEM, fixed-effect model; REM, random-effect model; BMI, body mass index.
Ethics Statement and Data Availability
This study was conducted in accordance with the Declaration of Helsinki. A written informed consent was obtained from all participants, and this study was approved by the ethics committee of Qilu Hospital and is numbered KYLL-2018 (KS)-135. All data generated or analyzed during this study are available via contacting the corresponding author.
Acknowledgments
This study was supported by Shandong University (Grant No. 26010132007081). We thank the participants and staff for their participation in the study. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Disclosure
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
1. Ozcan-Eksi EE, Eksi MS, Turgut VU, Canbolat C, Pamir MN. Reciprocal relationship between multifidus and psoas at L4-L5 level in women with low back pain. Br J Neurosurg. 2021;35:220–228. doi:10.1080/02688697.2020.1783434
2. Dario AB, Ferreira ML, Refshauge KM, Lima TS, Ordoñana JR, Ferreira PH. The relationship between obesity, low back pain, and lumbar disc degeneration when genetics and the environment are considered: a systematic review of twin studies. Spine J. 2015;15(5):1106–1117. doi:10.1016/j.spinee.2015.02.001
3. Wang F, Cai F, Shi R, Wang XH, Wu XT. Aging and age related stresses: a senescence mechanism of intervertebral disc degeneration. Osteoarthr Cartil. 2016;24(3):398–408. doi:10.1016/j.joca.2015.09.019
4. Toktas ZO, Eksi MS, Yilmaz B, et al. Association of collagen I, IX and vitamin D receptor gene polymorphisms with radiological severity of intervertebral disc degeneration in Southern European Ancestor. Eur Spine J. 2015;24(11):2432–2441. doi:10.1007/s00586-015-4206-5
5. Li YC, Zhu JJ, Gao CH, Peng BG. Vitamin D receptor (VDR) genetic polymorphisms associated with intervertebral disc degeneration. J Genet Genomics. 2015;42:135–140. doi:10.1016/j.jgg.2015.03.006
6. Cheung KM, Chan D, Karppinen J, et al. Association of the Taq I allele in vitamin D receptor with degenerative disc disease and disc bulge in a Chinese population. Spine. 2006;31(10):1143–1148. doi:10.1097/01.brs.0000216530.41838.d3
7. Vieira LA, Dos Santos AA, Peluso C, Barbosa CP, Bianco B, Rodrigues LMR. Influence of lifestyle characteristics and VDR polymorphisms as risk factors for intervertebral disc degeneration: a case-control study. Eur J Med Res. 2018;23(1):11. doi:10.1186/s40001-018-0309-x
8. Zawilla NH, Darweesh H, Mansour N, et al. Matrix metalloproteinase-3, vitamin D receptor gene polymorphisms, and occupational risk factors in lumbar disc degeneration. J Occup Rehabil. 2014;24(2):370–381. doi:10.1007/s10926-013-9472-7
9. Eskola PJ, Kjaer P, Daavittila IM, et al. Genetic risk factors of disc degeneration among 12–14-year-old Danish children: a population study. Int J Mol Epidemiol Genet. 2010;1(2):158–165.
10. Kawaguchi Y, Kanamori M, Ishihara H, Ohmori K, Matsui H, Kimura T. The association of lumbar disc disease with vitamin-D receptor gene polymorphism. J Bone Joint Surg Am. 2002;84(11):2022–2028. doi:10.2106/00004623-200211000-00018
11. Noponen-Hietala N, Kyllönen E, Männikkö M, et al. Sequence variations in the collagen IX and XI genes are associated with degenerative lumbar spinal stenosis. Ann Rheum Dis. 2003;62(12):1208–1214. doi:10.1136/ard.2003.008334
12. Withanage ND, Perera S, Peiris H, Athiththan LV. Serum 25-hydroxyvitamin D, serum calcium and vitamin D receptor (VDR) polymorphisms in a selected population with lumbar disc herniation-A case control study. PLoS One. 2018;13(10):e0205841. doi:10.1371/journal.pone.0205841
13. Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24(9):1291–1306. doi:10.1002/sim.2010
14. Ozcan-Eksi EE, Eksi MS, Akcal MA. Severe lumbar intervertebral disc degeneration is associated with Modic changes and fatty infiltration in the paraspinal muscles at all lumbar levels, except for L1-L2: a cross-sectional analysis of 50 symptomatic women and 50 age-matched symptomatic men. World Neurosurg. 2019;122:e1069–e1077. doi:10.1016/j.wneu.2018.10.229
15. Ekşi MŞ, Özcan-ekşi EE, Orhun Ö, Turgut VU, Pamir MN. Proposal for a new scoring system for spinal degeneration: mo-fi-disc. Clin Neurol Neurosurg. 2020;198:106120. doi:10.1016/j.clineuro.2020.106120
16. Ekşi MŞ, Özcan-ekşi EE, Özmen BB, et al. Lumbar intervertebral disc degeneration, end-plates and paraspinal muscle changes in children and adolescents with low-back pain. J Pediatr Orthop B. 2020. doi:10.1097/BPB.0000000000000833
17. Özcan-ekşi EE, Yayla A, Orhun Ö, Turgut VU, Arslan HN, Ekşi MŞ. Is the distribution pattern of Modic changes in vertebral end-plates associated with the severity of intervertebral disc degeneration?: a cross-sectional analysis of 527 caucasians. World Neurosurg. 2021;150:e298–e304. doi:10.1016/j.wneu.2021.02.128
18. Biczo A, Szita J, McCall I, Pal Varga P, Lazary A. Association of vitamin D receptor gene polymorphisms with disc degeneration. Eur Spine J. 2020;29(3):596–604. doi:10.1007/s00586-019-06215-7
19. Cauci S, Viganò M, de Girolamo L, et al. High levels of circulating type II collagen degradation marker (CTx-II) are associated with specific VDR polymorphisms in patients with adult vertebral osteochondrosis. Int J Mol Sci. 2017;18(10):2073–2088. doi:10.3390/ijms18102073
20. Cervin Serrano S, González Villareal D, Aguilar-Medina M, et al. Genetic polymorphisms of interleukin-1 alpha and the vitamin D receptor in Mexican mestizo patients with intervertebral disc degeneration. Int J Genomics. 2014;2014:302568. doi:10.1155/2014/302568
21. Colombini A, Brayda-Bruno M, Ferino L, et al. Gender differences in the VDR-FokI polymorphism and conventional non-genetic risk factors in association with lumbar spine pathologies in an Italian case-control study. Int J Mol Sci. 2015;16(2):3722–3739. doi:10.3390/ijms16023722
22. Kelempisioti A, Eskola PJ, Okuloff A, et al. Genetic susceptibility of intervertebral disc degeneration among young Finnish adults. BMC Med Genet. 2011;12:153. doi:10.1186/1471-2350-12-153
23. Kitis S, Coskun ZM, Tasdemir P, Tuncez E, Zamani AG, Acar A. Analysis of genetic polymorphisms associated with intervertebral disc degeneration. Cell Mol Biol. 2018;64(10):61–65. doi:10.14715/cmb/2018.64.10.10
24. Mashayekhi S, Saberi A, Salehi Z, Biazar G, Mehrdel R. VDR and GC gene polymorphisms modulate the risk of lumbar disc degeneration in Iran. Clin Neurol Neurosurg. 2018;165:67–71. doi:10.1016/j.clineuro.2017.12.024
25. Oishi Y, Shimizu K, Katoh T, et al. Lack of association between lumbar disc degeneration and osteophyte formation in elderly Japanese women with back pain. Bone. 2003;32(4):405–411. doi:10.1016/s8756-3282(03)00031-0
26. Ozdogan S, Yaltirik CK, Yilmaz SG, Kocak A, Isbir T. Association of rs2228570 polymorphism of vitamin D receptor gene with lumbar degenerative disc disease. Turk Neurosurg. 2019;29(2):159–163. doi:10.5137/1019-5149.JTN.22275-17.2
27. Yuan HY, Tang Y, Liang YX, et al. Matrix metalloproteinase-3 and vitamin d receptor genetic polymorphisms, and their interactions with occupational exposure in lumbar disc degeneration. J Occup Health. 2010;52(1):23–30. doi:10.1539/joh.l8149
28. Vieira LA, De Marchi PL, Dos Santos AA, et al. Analysis of FokI polymorphism of vitamin D receptor gene in intervertebral disc degeneration. Genet Test Mol Biomarkers. 2014;18(9):625–629. doi:10.1089/gtmb.2014.0030
29. Xu GH, Xu J, Zheng B, Yang SX, Lin XZ. Association of vitamin D receptor rs731236 gene polymorphisms with lumbar degenerative disc disease with Han nationality in Fujian (in Chinese). Chin J Bone Joint Inj. 2014;2014(9):882–884.
30. Chen WJ, Ye W, Ding Y, et al. Association of vitamin D receptor gene trui and rs2228570 polymorphisms with lumbar degenerative disease in Han nationality (in Chinese). Orthop J China. 2007;15(5):373–375.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.