Back to Journals » OncoTargets and Therapy » Volume 11
Association between WWOX and the risk of malignant tumor, especially among Asians: evidence from a meta-analysis
Authors Yuan H, Wu Y , Zhang J, Shi G, Liu D, He Y, Lu Z, Wu P, Jiang K, Miao Y
Received 20 September 2017
Accepted for publication 28 November 2017
Published 3 April 2018 Volume 2018:11 Pages 1801—1811
DOI https://doi.org/10.2147/OTT.S152140
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Samir Farghaly
Hao Yuan,1,2,* Yang Wu,1,2,* Jingjing Zhang,1,2 Guodong Shi,1,2 Dongfang Liu,1,2 Yuan He1,2 Zipeng Lu,1,2 Pengfei Wu,1,2 Kuirong Jiang,1,2 Yi Miao1,2
1Pancreas Center, Department of General Surgery, The First Affiliated Hospital of Nanjing Medical University, 2Pancreas Institute, Nanjing Medical University, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Purpose: Many studies have been carried out to examine whether there are associations between WWOX polymorphisms (rs3764340 C>G, rs12918952 G>A, and rs383362 G>T) and malignant tumor risk, but the results from these studies remained inconsistent and even controversial. In the present study, we performed a meta-analysis to evaluate the relationships comprehensively.
Methods: Published reports were searched in PubMed, Google Scholar, and Chinese National Knowledge Infrastructure databases. Eight eligible case–control studies were included in the final analysis. In the analysis, pooled odds ratios (ORs) with corresponding 95% CIs were calculated in five genetic models to assess the genetic risk. Egger’s regression and Begg’s funnel plots test were conducted to appraise the publication bias.
Results: We found that rs12918952 G>A and rs383362 G>T polymorphisms were not associated with the susceptibility of malignant tumor. However, a significant correlation was found between WWOX rs3764340 C>G and malignant tumor risk in three genetic models (CG vs CC: OR=1.31, 95% CI: 1.12–1.53, P=0.031; GG/CG vs CC: OR=1.31, 95% CI: 1.11–1.54, P=0.014; G vs C: OR=1.28, 95% CI: 1.09–1.50, P=0.009). Furthermore, when stratified by source of control, the results were significant especially in population-based control for rs3764340.
Conclusion: In general, our results first indicated that the rs3764340 C>G polymorphism in WWOX gene can increase the susceptibility of tumor, while the others cannot. However, large, well-designed epidemiological studies are required to verify our findings.
Keywords: malignant tumor, WWOX, meta-analysis, polymorphism, susceptibility
Introduction
Malignant tumor represents an enormous economic burden on society in both developing and developed countries. According to 32 GLOBOCAN estimates, approximately 14.1 million new cancer cases and 8.2 million deaths occurred in 2012 worldwide, especially in the developing countries.1 Cancer is a complex multifaceted disease that results from gene–environment interactions. Apart from the common factors such as smoking, obesity, and less physical activity, the genetic variations, including single nucleotide polymorphisms (SNPs), have been reported to be associated with malignant tumor susceptibility. The WWOX gene, located on chromosome 16q23.3-24.1, has been reported to play an anticancer role by regulating signaling pathways and cellular functions.2–4 In addition, both in vivo and in vitro studies have indicated that WWOX can induce apoptosis by interacting with p53 and p73.5–7 Low expression and abnormal transcription of WWOX has been found in several types of cancer, including lung cancer, prostate cancer, breast cancer, and oral cancer.8–13 Moreover, the low expression of this gene is correlated with higher tumor stages and worse prognosis in patients.14 In several studies, SNPs of WWOX gene have been identified as a potentially genetic risk factor in malignant tumors including lung cancer, gastric cancer, thyroid cancer, esophageal cancer, and osteosarcoma.15–20 However, other studies indicated that there were no significant associations.21,22 Because of relatively small sample sizes, these studies provided limited evidence and might have been underpowered to detect the overall effects. Therefore, we conducted this study to obtain a more precise estimation from all eligible case–control studies.
Methods
Primary search strategy
We systematically searched PubMed, Google Scholar, and Chinese National Knowledge Infrastructure comprehensively for all publications regarding the association between the WWOX polymorphisms and the malignant tumor risk (up to August 30, 2017), by using the combinations of the following keywords: WWOX, variants/polymorphism/SNP, and malignant tumor. Additional usable data were hand-searched from reference lists of original studies or review articles. Nevertheless, if several studies were performed on the same subjects, only the one with latest and/or largest sample size would be involved. Ethical approval was not necessary because this was a meta-analysis, including no direct handing of personal data or recruitment of subjects.
Inclusion and exclusion criteria
Studies involved had to satisfy the following inclusion criteria: case–control design was utilized; the diagnosis of the patients with cancer should be pathologically confirmed and the controls were verified as free from any cancer; and sufficient data for estimating an odds ratio (OR) with 95% CI was available. The major exclusion criteria were as follows: no obtainable genotype frequency data; duplicates of previous publication; and studies designed as a case–case or case-only study.
Data extraction
The identified studies were reviewed separately by two investigators (HY, YW) independently and carefully to determine whether an individual study was eligible for the analysis. These data were extracted from studies involved independently, and the disagreement was resolved by a discussion involving a senior investigator (KJ). All the following data were sought from each study and recorded in a standardized form: first author’s name; year of publication; ethnicity of each study population; source of controls; sample size; genotyping method; number of cases and controls; and frequencies of rs3764340, rs12918952, and rs383362 in cases and controls, respectively; and results of the Hardy–Weinberg equilibrium (HWE) test.
Statistical analysis
The pooled ORs with 95% CIs were applied to evaluate the strength of association between WWOX rs3764340 C>G, rs12918952 G>A, and rs383362 G>T polymorphisms and malignant tumor susceptibility. The fixed-effects model (the Mantel–Haenszel method) and the random-effects model (the DerSimonian–Laird method) were separately employed to pool the data.23 If existence of heterogeneity was detected, the random-effects model would be more appropriate. Sensitivity analysis was performed to assess the influence of individual studies on the pooled ORs, with the method of calculating the outcomes again by omitting one single study each time. After that, subgroup analyses based on the source of controls was further carried out to identify the potential association for each subgroup. Begg’s funnel plots and Egger’s linear regression method were taken to estimate the publication bias, and a P<0.05 was set as the significance threshold.24 HWE was checked by the goodness-of-fit chi-square test, and a P<0.05 was considered as a significantly selective bias.25
Stata software (version 12.0; StataCorp LP, College Station, TX, USA) was employed in the whole statistical analyses. P-values were all two-sided and regarded as statistically significant if less than 0.05.
Results
Studies characteristics
The flowchart of study exclusion and inclusion criteria with specific reasons is indicated in Figure 1. We identified 30 records, among which 14 articles appeared to be potentially eligible for inclusion and were retrieved in full texts. After full-text review, six articles were excluded due to no detailed data. Therefore, a total of eight case–control studies were ultimately included in the meta-analysis,15–22 and the details of each study were recorded in Table 1. Among them, eight articles containing 6,177 cases and 6,606 controls surveyed the association between rs3764340 C>G and malignant tumor risk.15–22 Four studies on the relation between rs12918952 G>A and malignant tumor risk included 2,447 cases and 3,249 controls,18,20–22 and four publications containing 2,950 cases and 3,872 controls explored the correlation of rs383362 G>T with malignant tumor risk.18,20–22 The genotyping methods used to detect the WWOX polymorphisms included sequencing, MassARRAY® System (Agena Bioscience, Inc., San Diego, CA, USA), polymerase chain reaction-restriction fragment polymorphism (PCR-RFLP), and TaqMan® (Thermo Fisher Scientific, Waltham, MA, USA). Furthermore, we divided them into population-based group or hospital-based group in all studies to distinguish between different sources of control group. For the WWOX rs3764340 polymorphism, the studies consisted of three population-based controls16,17,21 and five hospital-based controls.15,18–20,22 For rs12918952 G>A and rs383362 G>T, one population-based control21 and three hospital-based controls18,20,22 were included in these studies, respectively. We used subgroup analyses to explore the effects of different sources of control and genotyping methods on the associations of increased risk of tumor with the WWOX rs3764340 C>G, rs12918952 G>A, and rs383362 G>T polymorphisms. In addition, the studied tumor type included gastric cancer, lung cancer, oral cancer, osteosarcoma, thyroid carcinoma, hepatocellular carcinoma, and esophageal cancer.
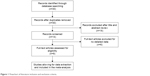  | Figure 1 Flowchart of literature inclusion and exclusion criteria. |
Quantitative synthesis results
The results for the association of WWOX rs3764340 C>G with malignant tumor risk are summarized in Table 2. Overall, neither the homozygous nor recessive genetic models found significant associations between rs3764340 C>G and malignant tumor risk (GG vs CC: OR=1.36, 95% CI: 0.90–2.04, P=0.392; GG vs CC/GC: OR=1.30, 95% CI: 0.88–1.93, P=0.451). The heterozygote and dominant genetic models witness obvious association between the SNPs and cancer risk (Figure 2) (CG vs CC: OR=1.31, 95% CI: 1.12–1.53, P=0.031; GG/CG vs CC: OR=1.31, 95% CI: 1.11–1.54, P=0.014). The allele analysis also found significant association (G vs C: OR=1.28, 95% CI: 1.09–1.50, P=0.009). For the different genotyping methods, rs3764340 C>G demonstrated increased risk of cancer both in the PCR-RFLP subgroup (CG vs CC: OR=1.46, 95% CI: 1.24–1.72, P=0.691; GG vs CC: OR=1.14, 95% CI: 0.53–2.45, P=0.239; GG/CG vs CC: OR=1.44, 95% CI: 1.23–1.68, P=0.532; GG vs CC/GC: OR=1.09, 95% CI: 0.51–2.32, P=0.248; G vs C: OR=1.38, 95% CI: 1.09–1.50, P=0.349) and in the TaqMan subgroup (CG vs CC: OR=1.32, 95% CI: 1.10–1.59, P=0.236; GG vs CC: OR=1.58, 95% CI: 0.94–2.66, P=0.566; GG/CG vs CC: OR=1.35, 95% CI: 1.10–1.66, P=0.158; GG vs CC/GC: OR=1.50, 95% CI: 0.90–2.52, P=0.651; G vs C: OR=1.34, 95% CI: 1.09–1.63, P=0.133). For the subgroup analysis according to the source of control, significant association between rs3764340 C>G and malignant tumor risk was found only in population-based group (CG vs CC: OR=1.33, 95% CI: 1.09–1.61, P=0.312; GG/CG vs CC: OR=1.35, 95% CI: 1.11–1.65, P=0.263; G vs C: OR=1.34, 95% CI: 1.11–1.63, P=0.236).
The results for the association of WWOX rs383362 with malignant tumor risk are summarized in Table 3. Overall, all genotypes were not significantly associated with the risk of malignant tumor (TG vs GG: OR=1.14, 95% CI: 0.89–1.45, P=0.008; TT vs GG: OR=1.04, 95% CI: 0.55–1.97, P=0.130; TT+GT vs GG: OR=1.13, 95% CI: 0.87–1.47, P=0.002; TT vs GT+GG: OR=1.02, 95% CI: 0.57–1.81, P=0.186). Similarly, no significant association was observed in the allele analysis (T allele vs G allele: OR=1.10, 95% CI: 0.86–1.41, P=0.002). In the subgroup analysis according to genotyping methods (Figure 3), the rs383362 G>T was associated with higher risk of malignant tumor in the TaqMan subgroup (TG vs GG: OR=1.35, 95% CI: 1.16–1.56, P=0.379; TT vs GG: OR=1.51, 95% CI: 1.03–2.22, P=0.758; TT+GT vs GG: OR=1.36, 95% CI: 1.18–1.57, P=0.364; TT vs GT+GG: OR=1.40, 95% CI: 0.95–2.05, P=0.811). The same result was found in the allele analysis (T allele vs G allele: OR=1.31, 95% CI: 1.16–1.49, P=0.370).
Table 4 shows the results for the association between rs12918952 G>A and cancer risk. Over all, no significant association was observed in any of the compared genetic models (AG vs GG: OR=1.06, 95% CI: 0.90–1.26, P=0.523; AA vs GG: OR=2.18, 95% CI: 0.62–0.768, P=0.216; AA+AG vs GG: OR=1.09, 95% CI: 0.92–1.29, P=0.38; AA vs AG+GG: OR=2.15, 95% CI: 0.62–7.49, P=0.223). Also, no significant association was revealed in the allele analysis (A allele vs G allele: OR=1.12, 95% CI: 0.92–1.35, P=0.254). However, a different result was found in the PCR-RFLP subgroup (Figure 4) (AA vs GG: OR=7.46, 95% CI: 1.17– 47.48, P=0.446; AA vs AG+GG: OR=7.31, 95% CI: 1.15–46.49, P=0.445) in the analysis according to different genotyping methods.
Heterogeneity test and sensitivity analysis
In most comparisons of WWOX rs3764340, rs383362, and rs12918952 polymorphisms, obvious heterogeneity was observed (figure not shown). We next performed a leave-one-out sensitivity analysis. The results of associations for WWOX rs3764340, rs383362, and rs12918952 polymorphisms in three important types of models are listed in Figure S1, which demonstrated stability and reliability of results for such associations.
Publication bias
We assessed the potential publication bias for three types of models (dominant model, heterozygous model, and allele model) by the Begg’s test and Egger’s test, and the results are shown in Table 5. No obvious symmetric distribution was found, indicating no evidence of significant publication bias. The overall results revealed that our results were statistically reliable.
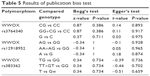  | Table 5 Results of publication bias test |
Discussion
WWOX, a protein with the WW domain, is conserved in all eukaryotes and has a significant role in cell signaling or regulation, such as regulating the protein degradation, mitosis, apoptosis, transcription, and RNA splicing process.26 Low or reduced expression of the WWOX protein was found in a variety of human malignancies, which may lead to the deregulation of these pathways. The silencing of the WWOX protein expression was observed very early in tumor development, which may result in the transformation of normal cells to malignant cells. The same result was found in the knock-out mice experiments.3 Based on the research of Maeda et al, low or reduced expression of WWOX was observed in gastric carcinoma cell lines (HSC-45, HSC-57, HSC-59, MKN-7, and MKN-74). Moreover, the gastric tumor tissue analyses showed that the loss of WWOX expression in 33% of cases was associated with poorer clinical findings.27 In addition, according to the studies of Dias et al, 91% of subjects with papillary thyroid carcinomas had a decreased expression of WWOX in the tumor tissue. Thus, it might indicate that the reduced expression of WWOX is the reason for cell proliferation, leading to further genomic instability.28
Polymorphisms have been identified as a powerful tool for predicting hereditary susceptibility of some complex diseases including malignant tumor.29 However, previous individual studies about the association between WWOX polymorphisms and malignant tumor risk were not only limited but also inconclusive. To our knowledge, this is the first comprehensive analysis researching the possible relationship of WWOX SNPs (rs3764340, rs383362, and rs12918952) with cancer susceptibility, which will be of benefit to provide new biomarkers for screening high-risk populations for malignant tumor and promoting the process of molecular-targeted therapy.
Rs3764340 C>G, which is located in exon 8, was associated with increased risk of multiple cancers.15–18,20 Based on the bioinformatics analysis, rs3764340 C>G is an exonic splicing enhancer which can promote the process of pre-mRNA splicing. Therefore, it is possible that this SNP may enhance the selective splicing in exon 8, leading to a high incidence of deletion of the exon.30 Another study also reported that the SNP may change the conformation of the WWOX protein.15 In this study, we found that WWOX 3764340 C>G was associated with risk of overall malignant tumor in the dominant model, heterozygous model, and allele model. In addition, in the subgroup analysis of different sources of controls, evidence of an association between the rs3764340 polymorphism and an increased cancer risk was only found in the population-based studies, but not in the hospital-based studies, which was probably caused by the fact that the controls recruited from hospital could not represent the general population well.
We did not find obvious association between SNP rs383362, rs12918952, and malignant tumor risk in any genetic comparison. In addition, only in the subgroup of TaqMan, WWOX rs38336 was constantly associated with increased tumor risk in all the compared genetic models. However, when it came to WWOX rs12918952, the association was only found in the subgroup of PCR-RFLP, which indicated that different genotyping methods might influence the results. It is worth noting that only four case–control studies were available for rs383362 and rs12918952 polymorphism analysis, and the sample size was not large enough. Therefore, the results should be interpreted with caution.
Some limitations of our meta-analysis should be noted. First, the number of studies was not sufficiently large, especially for subgroup analysis of WWOX rs383362 and rs12918952. Second, the languages of the publications were limited to English and Chinese. Finally, the lack of original data in some valuable research articles limited us to continue researching some potential interactions, such as age, sex, family history, environmental factors, and lifestyle. Consequently, it is required that a more precise analysis be performed if individual data were available.
Conclusion
The WWOX rs3764340 GG+CG and CG genotypes may increase cancer risk. However, rs383362 and rs12918952 polymorphisms may have no significant association with malignant tumor risk. Further large-scale and well-designed investigations including different cancers are required to verify the findings of this study.
Acknowledgments
I am profoundly grateful to my supervisor, Kuirong Jiang, whose expert advice has guided me through every step of my writing of this thesis. Thanks to Jie Yin and Dong Xu for their support in writing the article. Thanks to Kai Zhang for his kind help in data analysis.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Chang NS, Doherty J, Ensign A, et al. Molecular mechanisms underlying WOX1 activation during apoptotic and stress responses. Biochem Pharmacol. 2003;66(8):1347. | ||
Aqeilan RI, Croce CM. WWOX in biological control and tumorigenesis. J Cel Physiol. 2007;212(2):307. | ||
Salah Z, Aqeilan R, Huebner K. WWOX gene and gene product: tumor suppression through specific protein interactions. Future Oncol. 2010;6(2):249. | ||
Aqeilan RI, Pekarsky Y, Herrero JJ, et al. Functional association between WWOX tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci U S A. 2004;101(13):4401–4406. | ||
Zawackapankau J, Kostecka A, Sznarkowska A, et al. p73 tumor suppressor protein: a close relative of p53 not only in structure but also in anti-cancer approach? Cell Cycle. 2010;9(4):720. | ||
Chang NS, Doherty J, Ensign A, et al. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J Biol Chem. 2005;280(52):43100. | ||
Gourley C, Paige AJ, Taylor KJ, et al. WWOX mRNA expression profile in epithelial ovarian cancer supports the role of WWOX variant 1 as a tumour suppressor, although the role of variant 4 remains unclear. Int J Oncol. 2005;26(6):1681. | ||
Guler G, Uner A, Guler N, et al. The fragile genes FHIT and WWOX are inactivated coordinately in invasive breast carcinoma. Cancer. 2004;100(8):1605. | ||
Aqeilan RI, Kuroki T, Pekarsky Y, et al. Loss of WWOX expression in gastric carcinoma. Clin Cancer Res. 2004;10(9):3053. | ||
Nunez MI, Ludes-Meyers J, Abba MC, et al. Frequent loss of WWOX expression in breast cancer: correlation with estrogen receptor status. Breast Cancer Res Treat. 2005;89(2):99–105. | ||
Pimenta FJ, Gomes DA, Perdigão PF, et al. Characterization of the tumor suppressor gene WWOX in primary human oral squamous cell carcinomas. Int J Cancer. 2010;118(5):1154–1158. | ||
Qin HR, Iliopoulos D, Semba S, et al. A role for the WWOX gene in prostate cancer. Cancer Res. 2006;66(13):6477–6481. | ||
Nunez MI, Rosen DG, Ludes-Meyers JH, et al. WWOX protein expression varies among ovarian carcinoma histotypes and correlates with less favorable outcome. BMC Cancer. 2005;5(1):1–10. | ||
Cancemi L, Romei C, Bertocchi S, et al. Evidences that the polymorphism Pro-282-Ala within the tumor suppressor gene WWOX is a new risk factor for differentiated thyroid carcinoma. Int J Cancer. 2011;129(12):2816–2824. | ||
Guo W, Wang G, Dong Y, et al. Decreased expression of WWOX in the development of esophageal squamous cell carcinoma. Mol Carcinog. 2013;52(4):265. | ||
Guo W, Dong Z, Dong Y, et al. Genetic and epigenetic alterations of WWOX in the development of gastric cardia adenocarcinoma. Environ Mol Mutagen. 2013;54(2):112–123. | ||
Huang D, Qiu F, Yang L, et al. The polymorphisms and haplotypes of WWOX gene are associated with the risk of lung cancer in southern and eastern Chinese populations. Mol Carcinog. 2011;32(11):1299–1308. | ||
Wang X, Ma K, Chi L, et al. Combining telomerase reverse transcriptase genetic variant rs2736100 with epidemiologic factors in the prediction of lung cancer susceptibility. J Cancer. 2016;7(7):846. | ||
Zhang N, Jiang Z, Ren W, et al. Association of polymorphisms in WWOX gene with risk and outcome of osteosarcoma in a sample of the young Chinese population. Onco Targets Ther. 2016;9:807–813. | ||
Cheng HL, Liu YF, Su CW, et al. Functional genetic variant in the Kozak sequence of WW domain-containing oxidoreductase (WWOX) gene is associated with oral cancer risk. Oncotarget. 2016;7(43):69384–69396. | ||
Lee HL, Cheng HL, Liu YF, et al. Functional genetic variant of WW domain-containing oxidoreductase (WWOX) gene is associated with hepatocellular carcinoma risk. PLoS One. 2017;12(4):e0176141. | ||
Dersimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28(2):105. | ||
Egger M, Davey SG, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Sun WG, Thompson EA. Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics. 1992;48(2):361–372. | ||
Lu PJ, Zhou XZ, Shen M, et al. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283(5406):1325. | ||
Maeda N, Semba S, Nakayama S, et al. Loss of WW domain-containing oxidoreductase expression in the progression and development of gastric carcinoma: clinical and histopathologic correlations. Virchows Arch. 2010;457(4):423–432. | ||
Dias EP, Pimenta FJ, Sarquis MS, et al. Association between decreased WWOX protein expression and thyroid cancer development. Thyroid. 2007;17(11):1055. | ||
Hirschhorn JN, Lohmueller K, Byrne E, et al. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45. | ||
Zhou Y, Xu Y, Zhang Z. Deletion and mutation of WWOX exons 6–8 in human non-small cell lung cancer. J Huazhong Univ Sci Technolog Med Sci. 2005;25(2):162–165. |
Supplementary materials
References
Wang X, Ma K, Chi L, et al. Combining telomerase reverse transcriptase genetic variant rs2736100 with epidemiologicfactors in the prediction of lung cancer susceptibility. J Cancer. 2016;7(7):846. | ||
Cheng HL, Liu YF, Su CW, et al. Functional genetic variant in the Kozak sequence of WW domain-containing oxidoreductase (WWOX) gene is associated with oral cancer risk. Oncotarget. 2016;7(43):69384–69396. | ||
Huang D, Qiu F, Yang L, et al. The polymorphisms and haplotypes of WWOX gene are associated with the risk of lung cancer in southern and eastern Chinese populations. Mol Carcinog. 2011;32(11):1299–1308. | ||
Cancemi L, Romei C, Bertocchi S, et al. Evidences that the polymorphism Pro-282-Ala within the tumor suppressor gene WWOX is a new risk factor for differentiated thyroid carcinoma. Int J Cancer. 2011;129(12):2816–2824. | ||
Guo W, Wang G, Dong Y, et al. Decreased expression of WWOX in the development of esophageal squamous cell carcinoma. Mol Carcinog. 2013;52(4):265. | ||
Guo W, Dong Z, Dong Y, et al. Genetic and epigenetic alterations of WWOX in the development of gastric cardia adenocarcinoma. Environ Mol Mutagen. 2013;54(2):112–123. | ||
Zhng N, Jiang Z, Ren W, et al. Association of polymorphisms in WWOX gene with risk and outcome of osteosarcoma in a sample of the young Chinese population. Onco Targets Ther. 2016;9:807–813. | ||
Lee HL, Cheng HL, Liu YF, et al. Functional genetic variant of WW domain-containing oxidoreductase (WWOX) gene is associated with hepatocellular carcinoma risk. PLoS One. 2017;12(4):e0176141. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.








