Back to Journals » Infection and Drug Resistance » Volume 13
Association Between the Phenotype and Genotype of Isoniazid Resistance Among Mycobacterium tuberculosis Isolates in Thailand
Authors Charoenpak R , Santimaleeworagun W, Suwanpimolkul G , Manosuthi W, Kongsanan P , Petsong S , Puttilerpong C
Received 13 December 2019
Accepted for publication 31 January 2020
Published 24 February 2020 Volume 2020:13 Pages 627—634
DOI https://doi.org/10.2147/IDR.S242261
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Ratchanu Charoenpak, 1 Wichai Santimaleeworagun, 2 Gompol Suwanpimolkul, 3–5 Weerawat Manosuthi, 6 Paweena Kongsanan, 6 Suthidee Petsong, 7 Chankit Puttilerpong 8
1College of Pharmacotherapy Thailand, Nonthaburi, Thailand; 2Department of Pharmacy, Faculty of Pharmacy, Silpakorn University, Nakhon Pathom, Thailand; 3Department of Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand; 4Tuberculosis Research Unit, Chulalongkorn University, Bangkok, Thailand; 5Emerging Infectious Diseases Clinical Center, Thai Red Cross, Bangkok, Thailand; 6Bamrasnaradura Infectious Diseases Institute, Ministry of Public Health, Nonthaburi, Thailand; 7Department of Microbiology, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand; 8Department of Pharmacy Practice, Faculty of Pharmaceutical Sciences, Chulalongkorn University, Bangkok, Thailand
Correspondence: Chankit Puttilerpong
Department of Pharmacy Practice, Faculty of Pharmaceutical Sciences, Chulalongkorn University, Phayathai Road, Patumwan, Bangkok, Thailand
Tel|Fax +66 2 218 8403
Email [email protected]
Purpose: The emergence of isoniazid-resistant tuberculosis (HR-TB) is a global public health problem, causing treatment failure and high mortality rates. This study aimed to determine the minimal inhibitory concentration (MIC) of isoniazid and detect the gene mutation in HR-TB and any association between the level of isoniazid resistance and gene mutation.
Methods: We collected 74 clinical HR-TB isolates from two tertiary-care centers in Thailand. MICs were established using broth macrodilution. A line probe assay (LPA) was used to detect gene mutations that confer resistance to isoniazid, rifampicin, aminoglycosides, and fluoroquinolones.
Results: Sixty-one (82.4%) isolates were monoresistant to isoniazid and 44 (72.1%) were highly resistant to isoniazid. From the clinical isolates, the range of isoniazid MICs was 0.4– 16 μg/mL. The katG S315T gene mutation was the prominent mutation in both isoniazid-monoresistant TB (70.5%) and multidrug-resistant TB (72.7%) isolates. The positive predictive value (PPV) of katG was 100% in detecting high levels of isoniazid resistance. The PPV of the inhA mutation was 93.8% in detecting low levels of isoniazid resistance. Five isolates (6.8%) exhibited low-level phenotypic resistance, whereas an LPA failed to detect an isoniazid gene mutation. Our study found one HR-TB isolate with a gyrA fluoroquinolone-resistant gene mutation.
Conclusion: Most HR-TB isolates had high isoniazid-resistance levels associated with the katG gene mutation. High-dose isoniazid should be used with caution in patients with HR-TB. Early detection of drug resistance by genotypic assay can help determine an appropriate regimen.
Keywords: tuberculosis, isoniazid, minimal inhibitory concentration, line probe assay, gene mutation
Introduction
Tuberculosis (TB) is an important cause of death worldwide and a public health crisis, even though it is preventable, treatable, and curable. Thailand is one of the 14 high-burden countries for TB, multidrug-resistant TB (MDR-TB), and TB-HIV coinfection, according to the World Health Organization (WHO). The 2018 annual report for TB in Thailand estimated 106,000 cases of TB and 11,000 TB-related deaths.1
Early detection of drug-resistant TB is important to effectively treat and prevent transmission and reduce mortality.2 According to the 2019 WHO global report, new cases of isoniazid-resistant tuberculosis (HR-TB) (7.2%) were more prevalent than MDR-TB (3.4%). The previously treated TB cases, a prevalence of HR-TB (11.6%) was less than that of MDR-TB/rifampicin-resistant TB (18%).1 In 1997–2013, a report on national anti-TB drug resistance in Thailand found HR-TB in 9.5%–12.2% new cases and 30%–44% retreatment cases.3
Isoniazid resistance in Mycobacterium tuberculosis (MTB) is associated with mutations of katG, inhA, ahpC promoter, and fabG1 genes.4 However, katG is the major isoniazid-resistant gene mutation found in 40%–95% of the clinical TB isolates,5–7 and katG S315T is the most frequently mutated gene found in isolates with a high level of isoniazid resistance.8 katG gene encodes catalase/peroxidase enzymes that activate isoniazid to become an active drug. Active isoniazid disrupts the mycolic acid biosynthesis by inhibiting inhA, the NADH-dependent enoyl-ACP reductase enzyme encoded by inhA gene.4 Another mechanism that causes low levels of isoniazid resistance is the inhA mutation, which is found in 20%–42% of the clinical TB isolates, and results in overexpression of isoniazid’s target and tends to increase the minimal inhibitory concentration (MIC).5 It also confers resistance to second-line drugs (ethionamide and prothionamide) as they share the same target site.9,10 WHO treatment guidelines, 2019, recommend treating patients with confirmed HR-TB with rifampicin, ethambutol, pyrazinamide, and levofloxacin (6REZ-Lfx) for 6 months.11 Rapid molecular tests for susceptibility to first- and second-line agents are recommended to help physicians in choosing an appropriate treatment regimen.11
Data from pharmacokinetics/pharmacodynamics (PK/PD) and in vitro microbiological data can help optimize the dosage of treatment drugs in a drug-resistant era. According to in vitro data, patients with TB or MDR-TB with a low level of resistance to isoniazid (MIC, 0.1–0.4 μg/mL in liquid medium or 0.2–1 μg/mL in solid medium) should be treated with isoniazid at a dosage of 15–20 mg/kg/day.12,13
In this study, we determined the MIC distribution of isoniazid by using broth macrodilution method (Bactec mycobacteria growth indicator tube [MGIT] 960 TB system) and assessed the gene mutations of clinical isolates for isoniazid, rifampicin, fluoroquinolones, and aminoglycosides by using a line probe assay (LPA). We also investigated the association between the level of isoniazid resistance and gene mutation.
Materials and Methods
Mycobacterium tuberculosis Isolates
Between January 2017 and April 2019, 98 clinical isolates were obtained from the Department of Microbiology, Faculty of Medicine, Chulalongkorn University, Bangkok, and the Microbiology Laboratory, of the Bamrasnaradura Infectious Diseases Institute, Nonthaburi, Thailand. Isoniazid resistance was confirmed through standard phenotypic drug-susceptibility test (DST) by using a Bactec MGIT 960 SIRE Kit. Of the isolates, 24 were excluded for absence of growth in the Löwenstein–Jansen (LJ) medium, contamination, or susceptibility to isoniazid on repeat DST. Therefore, 74 clinical isolates were analyzed: 59 (79.7%) from patients with pulmonary TB and 15 (20.3%) from patients with extrapulmonary TB. According to the MGIT SIRE results, 43 isolates (58.1%) were monoresistant to isoniazid, 18 (24.3%) were poly-resistant, and 13 (17.6%) were MDR. Of the poly-resistant isolates, 2 were resistant to isoniazid and ethambutol and 16 to isoniazid and streptomycin. Of the MDR isolates, 7 were resistant to isoniazid and rifampicin; 5 were resistant to isoniazid, rifampicin, and streptomycin; and 1 was resistant to isoniazid, rifampin, and ethambutol.
Each isolate was stored in MGIT broth until MIC analysis. The isolates were subcultured in an LJ medium and purity was ascertained by gross colonial morphology and LPA.
This study was approved by the institutional review board (IRB) of the Faculty of Medicine, Chulalongkorn University, Bangkok (IRB No. 046/62) and Bamrasnaradura Infectious Diseases Institute, Nonthaburi (IRB No. R005h/62_ExPD), Thailand. Written informed consent was waived for this study as we used archived MTB isolates.
Phenotypic Drug-Susceptibility Test
Each isolate was grown on LJ slants at 37°C for 15 days and then tested for MIC. The concentrations of isoniazid (Sigma-Aldrich, St Louis, MO, USA) used in this study were 0.1, 0.4, 1, 2, 4, 8, 16, and 32 μg/mL in Bactec MGIT TB broth with BD BBL Middlebrook Oleic Albumin Dextrose Catalase Enrichment (Becton, Dickinson and company, Sparks, MD, USA). A Bactec MGIT 960 TB system using the EpiCenter software interpreted the results, according to the manufacturer’s instructions. Isoniazid resistance was determined by the Clinical and Laboratory Standards Institute as follows: low- and high-level isoniazid resistance in liquid media had to have an MIC of 0.1–0.4 and >0.4 μg/mL, respectively.14 A pan-susceptible MTB routine QC strain was used as quality control.
Genotypic Drug-Susceptibility Test
Geno Type MTBDRplus version 2.0 (Hain Lifescience, Nehren, Germany) was used to detect mutations in katG (S315T), inhA (C-15T, A-16G, T-8C, and T-8A), and rifampicin (rpoB codon 505–533) genes, whereas MTBDRsl version 2.0 (Hain Lifescience, Nehren, Germany) was used to detect fluoroquinolone-resistance mutations (gyrA [G88A/C, A90V, S91P, D94A, D94N/Y, D94G, and D94H], and gyrB [N538D and E540V]) and aminoglycosides/cyclic peptides injectable antibiotic-resistant mutations (rrs [A1401G, C1402T, and G1484T] and eis [G-37T, C-14T, C-12T, G-10A, and C-2A]). Extraction, amplification, and hybridization of DNA were performed according to the manufacturer’s instructions.
WHO’s definitions15 for various anti-TB drug resistances were used. Mono-resistance indicated resistance to only one drug from the first-line anti-TB drug group. Poly-resistance indicated resistance to more than one first-line anti-TB drug, other than both isoniazid and rifampicin. MDR indicated resistance to isoniazid and rifampicin. Pre-XDR TB indicated MDR-TB with additional resistance to any fluoroquinolone or at least one drug from the second-line injectable drugs (capreomycin, kanamycin, or amikacin). Extensive drug resistance (XDR-TB) indicated MDR-TB and resistance to any fluoroquinolone and at least one drug from the second-line injectable drugs.
Data Analysis
Descriptive statistics such as frequencies and percentages were used to describe MIC and gene mutation data. All data were analyzed using IBM SPSS Statistics for Windows, Version 22.0.
Results
Of the 74 clinical HR-TB isolates analyzed in this study, 45 (60.8%) were obtained from the Department of Microbiology, Faculty of Medicine, Chulalongkorn University, Bangkok, and 29 (39.2%) were obtained from the Microbiology Laboratory at the Bamrasnaradura Infectious Diseases Institute, Nonthaburi. Of the included isolates, 61 (82.4%) were monoresistant to isoniazid, 11 (14.9%) were multidrug-resistant, 1 (1.4%) was poly-resistant, and 1 (1.4%) was pre-XDR. Genotypic drug resistance and the level of isoniazid resistance are shown in Figure 1.
 |
Figure 1 Genotypic drug resistance and level of resistance to isoniazid. |
The isoniazid MIC range was 0.4–16 μg/mL, whereas MIC50 and MIC90 were 2 and 4 μg/mL, respectively. A high level of resistance to isoniazid was exhibited by 54 (73%) isolates, of which 51 had katG S315T mutation, 2 had katG deletion mutation, and 1 had inhA mutation, with the katG deletion mutation having the highest MIC. Fifteen isolates (20.3%) had inhA mutation, which confers low-level resistance to isoniazid. Although isoniazid gene mutations were undetectable in 5 (6.8%) isolates, they exhibited low levels of resistance to isoniazid. Isoniazid MIC distribution and gene mutations of MTB isolates are summarized in Figure 2.
 |
Figure 2 Isoniazid MIC distribution and gene mutations of Mycobacterium tuberculosis. |
Phenotypic DST was used as a reference standard. katG (katG315T and katG deletion) and inhA gene mutations were detected by LPA, and they were associated with high and low levels of resistance to isoniazid, respectively. Isolates with katG mutation exhibited high-level resistance to isoniazid, with sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of 98.2%, 100%, 100%, and 93.8%, respectively. Isolates with inhA mutation exhibited low-level resistance to isoniazid, with sensitivity, specificity, PPV, and NPV of 100%, 98.2%, 93.8%, and 100%, respectively. Performance characteristics of these gene mutations and their levels of resistance are detailed in Table 1.
 |
Table 1 Performance Characteristics of the Isoniazid Gene Mutation and the Levels of Isoniazid Resistance in Clinical Isolates (N = 69)a |
Seventy isolates (94.6%) were detected to have drug-resistant gene mutation by using LPA. katG S315T (68.9%), inhA C-15T (21.6%), and katG deletion (2.7%) mutations were associated with isoniazid resistance. All isoniazid-resistant isolates had a single gene mutation. Twelve (16.2%) isolates had a rifampicin-resistant mutation, with the most prevalent of them being rpoB (S531L), which was identified in 6 MDR-TB isolates. The results of rifampicin resistance differed between phenotype and genotype tests. Thirteen isolates had phenotypic resistance and 12 had genotypic resistance (Additional details are provided in the Supplemental table) Mutation in the gyrA (D94G) gene can affect the encoding of fluoroquinolones. Only one isolate had this mutation, and its phenotypic resistance to isoniazid was low. The pre-XDR isolate had a mutation in the rrs (A1401G) gene, which affected the encoding of aminoglycosides. Gene mutations of MTB are presented in Table 2.
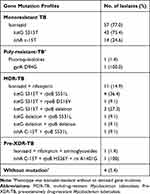 |
Table 2 Gene Mutation Profiles of MTB Isolates (N = 74) |
Discussion
This is the first study to demonstrate an association between the phenotype and genotype of numerous HR-TB isolates in Thailand. Antimicrobial susceptibility test endorsed by WHO was used in this study. Most clinical isolates had both high-level resistant HR-TB and the katG S315T gene mutation. A previous report found a high prevalence of the katG S315T gene mutation among HR-TB/MDR-TB/XDR-TB strains (58.7%–68.6%) in Thailand.16,17 The prevalence of katG S315T varied according to the geographic region: Southeast Asia (78.4%),18 Vietnam (85.3%),19 Myanmar (61.2%),20 Taiwan (29.3%),6 Japan (25%),10 India (67.6%),21 Sri Lanka (67.8%),22 Romania (52.8%),23 Cameroon (64%),24 and the United States (38%).8
The different ranges of MIC found for each mutation gene can be explained by gene variation. For example, in this study, katG S315T and inhA C-15T genes had MIC ranges of 1–4 and 0.4–4 μg/mL, respectively, whereas the katG deletion had an MIC of 1 and 16 μg/mL. In another study,25 which used the MGIT 960 TB system, the MIC was higher than that in our study; the MIC ranges for katG S315T, inhA promoter, inhA promoter with katG loss of function, and katG S315T with inhA promoter were 4–16, 0.25–2, 4 to >64, and 8–64 μg/mL, respectively. Previous studies had more than one gene mutations exhibiting a higher MIC range.5,18,25,26 However, in our study, we detected only a single gene mutation.
The discordance in the DSTs between genotype and phenotype can contribute to under-treatment of drug-resistant tuberculosis, which can in turn lead to resistant strains.2 We found that 6.8% of these isolates had low levels of isoniazid resistance, and the mutation gene could not be detected. Because of limitations in detecting isoniazid gene mutations, we could only detect katG and inhA mutations using LPA. Isoniazid gene mutations should be confirmed by whole-genome sequencing, which can detect uncommon isoniazid-resistance mutations such as katG S315N/G, ahpC promoter, or fabG1.26 Recent studies have shown that these uncommon gene mutations may be susceptible or have low-/high-level resistance to isoniazid.5,26 WHO recommends that DSTs of both phenotypes and genotypes should be used for all isolates.
The detection of gene mutations may help predict the level of isoniazid resistance and help physicians select the most appropriate treatment regimen for patients. Detection of the katG S315T gene was associated with high-level isoniazid resistance; in our study, the PPV was 100% and the NPV was 93.8%. Previous studies have shown that 61.7%–100% of the mutations occurred at amino acid position 315 in the katG gene, especially katG S315T, in HR-TB isolates with a high level of resistance to isoniazid.5,8,10,27 In our study, the PPV was 93.8% and the NPV was 100% for the inhA C-15T gene mutation, which is associated with low-level isoniazid resistance. We found that 1 out of 16 isolates had the inhA C-15T gene, corroborating the findings by Lempens et al5 who reported that 5.8% of the isolates included had the inhA C-15T gene mutation and a high level of isoniazid resistance. Mutation of the inhA gene does not always indicate low resistance levels.
Only 1.4% of HR-TB isolates (a patient who received retreatment was exposed to levofloxacin and had poor adherence) had a fluoroquinolone-resistant gene mutation. This agreed with a recent study conducted in Thailand that found a low prevalence of fluoroquinolone resistance in non-MDR-TB isolates resistant to ofloxacin (5.1%), levofloxacin (3.4%), and moxifloxacin (1.7%) by the phenotype method.28 While physicians wait for the DST results to confirm the phenotype of the TB, according to WHO 201911 and National Tuberculosis Control Programme Guideline, Thailand 2018,29 patients with HR-TB having severe symptoms and immune deficiency, TB meningitis, TB in the bone and joint, or disseminated TB should be treated with 6RZE-Lfx regimen. However, if a patient cannot tolerate fluoroquinolones or is pregnant or breastfeeding, a 6RZE regimen could be used. The mortality rate of isoniazid-monoresistant TB is higher than pan-susceptible TB when patients are treated with the standard first-line regimen.2,30
Zuur et al13 proposed using high-dose isoniazid (1200 mg/day) among patients whose isolate had an MIC of ≤0.5 μg/mL. In our study, approximately 27.0% of the HR-TB population had an MIC of ≤0.4 μg/mL. Before administering high doses of isoniazid, we recommend that NAT2 gene polymorphism and toxicity should be evaluated. Anti-TB drugs are well known to induce hepatitis-related dose-dependent toxicity. We therefore caution against the use of short-course, high-dose isoniazid to treat MDR-TB. Additionally, the prevalence of high levels of isoniazid resistance was 83.3%, making it difficult to achieve the target PK/PD.
Our study has some limitations. The findings are not representative of HR-TB of Thailand because of inclusion of only two centers and the small sample size. A multicenter trial with a larger sample size is needed to confirm our results. Although LPA is frequently used in clinical practice to detect resistance to both first- and second-line anti-TB drugs, it can only detect common mutations because it uses specific probes. Whole-genome sequencing can overcome this problem, but cost-efficacy assessment and additional studies are required for this option. In addition, some of the isolates were not the first specimen collected at the time of diagnosis. It is therefore difficult to classify the prevalence of primary drug resistance or acquired resistance. However, drug resistance in MTB isolates occurs at a low frequency because of spontaneous chromosomal mutations. Improper use of the regimen and patient nonadherence can contribute to acquired resistance. Early detection and improved adherence may prevent a drug-resistance crisis. In the future, we will examine the clinical outcomes of this population.
Conclusion
We found an association between phenotype and isoniazid-resistant gene mutation in HR-TB clinical isolates. All katG gene mutations conferred a high level of resistance to isoniazid. From the MIC data, a high-dose of isoniazid may not be appropriate for patients with HR-TB. The prevalence of fluoroquinolone drug resistance was low in HR-TB, indicating that 6RZE-Lfx can be used to treat HR-TB in Thailand.
Acknowledgments
The authors express their gratitude to the Doctor Kasem Pangsrivongse Foundation and the Faculty of Pharmaceutical Sciences, Chulalongkorn University, for providing research fund (Grant Number Phar2563-RG005) to Dr. Chankit Puttilerpong. Funders had no role in study design, data collection, analysis of data, or writing of the manuscript.
Disclosure
The authors declare no conflict of interest.
References
1. World Health Organization. Global Tuberculosis Report 2019. Geneva, Switzerland: WHO; 2019. Available from:https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714-eng.pdf?ua=1.
2. Zurcher K, Ballif M, Fenner L, et al. Drug susceptibility testing and mortality in patients treated for tuberculosis in high-burden countries: a multicentre cohort study. Lancet Infect Dis. 2019;19(3):298–307. doi:10.1016/S1473-3099(18)30673-X
3. Reechaipichitkul W, Nateniyom S, Pungrassami P, eds. Guidelines for Programmatic Management of Drug-Resistant Tuberculosis. Bangkok, Thai: Bureau of Tuberculosis Department of Disease Control; 2015. Available from:https://www.tbthailand.org/download/DR-TB%20Proof%2023%20FINAL.pdf.
4. Unissa AN, Subbian S, Hanna LE, Selvakumar N. Overview on mechanisms of isoniazid action and resistance in Mycobacterium tuberculosis. Infect Genet Evol. 2016;45:476–492. doi:10.1016/j.meegid.2016.09.004
5. Lempens P, Meehan CJ, Vandelannoote K, et al. Isoniazid resistance levels of Mycobacterium tuberculosis can largely be predicted by high-confidence resistance-conferring mutations. Sci Rep. 2018;8(1):3246. doi:10.1038/s41598-018-21378-x
6. Tseng ST, Tai CH, Li CR, Lin CF, Shi ZY. The mutations of katG and inhA genes of isoniazid-resistant Mycobacterium tuberculosis isolates in Taiwan. J Microbiol Immunol Infect. 2015;48(3):249–255. doi:10.1016/j.jmii.2013.08.018
7. Jabbar A, Phelan JE, de Sessions PF, et al. Whole genome sequencing of drug resistant Mycobacterium tuberculosis isolates from a high burden tuberculosis region of North West Pakistan. Sci Rep. 2019;9(1):14996. doi:10.1038/s41598-019-51562-6
8. Dantes R, Metcalfe J, Kim E, et al. Impact of isoniazid resistance-conferring mutations on the clinical presentation of isoniazid monoresistant tuberculosis. PLoS One. 2012;7(5):e37956. doi:10.1371/journal.pone.0037956
9. Vilcheze C, Jacobs WJ. Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: genes, mutations, and causalities. Microbiol Spectr. 2014;2(4):431–453. doi:10.1128/microbiolspec.MGM2-0014-2013
10. Abe C, Kobayashi I, Mitarai S, et al. Biological and molecular characteristics of Mycobacterium tuberculosis clinical isolates with low-level resistance to isoniazid in Japan. J Clin Microbiol. 2008;46(7):2263–2268. doi:10.1128/JCM.00561-08
11. World Health Organization. WHO Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment. Geneva, Switzerland: WHO; 2019. Available from:https://apps.who.int/iris/bitstream/handle/10665/311389/9789241550529-eng.pdf?ua=1.
12. Katiyar SK, Bihari S, Prakash S, Mamtani M, Kulkarni H. A randomised controlled trial of high-dose isoniazid adjuvant therapy for multidrug-resistant tuberculosis. Int J Tuberc Lung Dis. 2008;12(2):139–145.
13. Zuur MA, Pasipanodya JG, van Soolingen D, van der Werf TS, Gumbo T, Alffenaar JC. Intermediate susceptibility dose-dependent breakpoints for high-dose rifampin, isoniazid, and pyrazinamide treatment in multidrug-resistant tuberculosis programs. Clin Infect Dis. 2018;67(11):1743–1749. doi:10.1093/cid/ciy346
14. Clinical and Laboratory Standards Institute. Susceptibility testing of mycobacteria, norcardia spp., and other arerobic actinomycetes. In: CLSI Standard M24.
15. World Health Organization. Companion Handbook to the WHO Guidelines for the Programmatic Management of Drug-Resistant Tuberculosis. Geneva, Switzerland: WHO; 2014. Available from:https://apps.who.int/iris/bitstream/handle/10665/130918/9789241548809_eng.pdf?sequence=1.
16. Jaksuwan R, Tharavichikul P, Patumanond J, et al. Genotypic distribution of multidrug-resistant and extensively drug-resistant tuberculosis in northern Thailand. Infect Drug Resist. 2017;10:167–174. doi:10.2147/IDR.S130203
17. Faksri K, Kaewprasert O, Ong RT, et al. Comparisons of whole-genome sequencing and phenotypic drug susceptibility testing for Mycobacterium tuberculosis causing MDR-TB and XDR-TB in Thailand. Int J Antimicrob Agents. 2019;54(2):109–116. doi:10.1016/j.ijantimicag.2019.04.004
18. Seifert M, Catanzaro D, Catanzaro A, Rodwell TC. Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: a systematic review. PLoS One. 2015;10(3):e0119628. doi:10.1371/journal.pone.0119628
19. Hang NTL, Hijikata M, Maeda S, et al. Whole genome sequencing, analyses of drug resistance-conferring mutations, and correlation with transmission of Mycobacterium tuberculosis carrying katG-S315T in Hanoi, Vietnam. Sci Rep. 2019;9(1):15354. doi:10.1038/s41598-019-51812-7
20. Aye KS, Nakajima C, Yamaguchi T, et al. Genotypic characterization of multi-drug-resistant Mycobacterium tuberculosis isolates in Myanmar. J Infect Chemother. 2016;22(3):174–179. doi:10.1016/j.jiac.2015.12.009
21. Munir A, Kumar N, Ramalingam SB, et al. Identification and characterization of genetic determinants of isoniazid and rifampicin resistance in Mycobacterium tuberculosis in Southern India. Sci Rep. 2019;9(1):10283. doi:10.1038/s41598-019-46756-x
22. Karunaratne G, Wijesundera SS, Vidanagama D, Adikaram CP, Perera J. Significance of coexisting mutations on determination of the degree of isoniazid resistance in Mycobacterium tuberculosis strains. Microb Drug Resist. 2018;24(6):844–851. doi:10.1089/mdr.2017.0330
23. Ruesen C, Riza AL, Florescu A, et al. Linking minimum inhibitory concentrations to whole genome sequence-predicted drug resistance in Mycobacterium tuberculosis strains from Romania. Sci Rep. 2018;8(1):9676. doi:10.1038/s41598-018-27962-5
24. Abanda NN, Djieugoue JY, Lim E, et al. Diagnostic accuracy and usefulness of the genotype MTBDRplus assay in diagnosing multidrug-resistant tuberculosis in Cameroon? A cross-sectional study. BMC Infect Dis. 2017;17(1):379. doi:10.1186/s12879-017-2489-3
25. Ghodousi A, Tagliani E, Karunaratne E, et al. Isoniazid resistance in Mycobacterium tuberculosis is a heterogeneous phenotype composed of overlapping MIC distributions with different underlying resistance mechanisms. Antimicrob Agents Chemother. 2019;63(7). doi:10.1128/AAC.00092-19.
26. Kandler JL, Mercante AD, Dalton TL, et al. Validation of novel Mycobacterium tuberculosis isoniazid resistance mutations not detectable by common molecular tests. Antimicrob Agents Chemother. 2018;62(10). doi:10.1128/AAC.00974-18.
27. van Soolingen D, de Haas PEW, van Doorn HR, Kuijper E, Rinder H, Borgdorff MW. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in the Netherlands. J Infect Dis. 2000;182(6):1788–1790. doi:10.1086/317598
28. Leechawengwongs M, Prammananan T, Jaitrong S, et al. In vitro activity and MIC of sitafloxacin against multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis isolated in Thailand. Antimicrob Agents Chemother. 2017;62(1):e00825–17. doi: 10.1128/AAC.00825-17
29. Bureau of tuberculosis, Department of disease control. National Tuberculosis Control Programme Guideline, Thailand, 2018.
30. Anuwatnonthakate A, Whitehead SJ, Varma JK, et al. Effect of mycobacterial drug resistance patterns on patients’ survival: a cohort study in Thailand. Glob J Health Sci. 2013;5(6):60–72. doi:10.5539/gjhs.v5n6p60
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
