Back to Journals » OncoTargets and Therapy » Volume 11
Association between KIF1B (rs17401966) polymorphism and hepatocellular carcinoma susceptibility: a meta-analysis
Authors Zhang Y , Zeng X, Lu H, Ji H, Lu L, Liu P, Hong R, Li Y
Received 11 January 2018
Accepted for publication 8 April 2018
Published 29 May 2018 Volume 2018:11 Pages 3225—3235
DOI https://doi.org/10.2147/OTT.S162205
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Samir Farghaly
Ya-fei Zhang,1 Xian-ling Zeng,2 Hong-wei Lu,1 Hong Ji,1 Le Lu,1 Peng-di Liu,1 Ruo-feng Hong,1 Yi-ming Li1
1Department of General Surgery, The Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi, China; 2Department of Obstetrics and Gynecology, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi, China
Introduction: The results of the earlier published studies on the association between KIF1B (rs17401966) polymorphism and hepatocellular carcinoma (HCC) risk are inconclusive. Hence, we performed this meta-analysis to evaluate the relationship between KIF1B (rs17401966) polymorphism and HCC risk.
Methods: Databases including PubMed, Web of Science and the Cochrane Library and bibliographies of relevant papers were screened to identify relevant studies published up to March 25, 2018. Pooled ORs and 95% CIs were calculated to evaluate the association. The subgroup analysis was conducted based on ethnicity, age, region and environment. A total of 19 studies from 11 eligible articles published from 2010 to 2016, with 8,741 cases and 10,812 controls, were included.
Results: The pooled results indicated that the association between KIF1B (rs17401966) polymorphism and the decreased HCC risk was significant. Subgroup analysis stratified by ethnicity showed the same association in Chinese, but not in non-Chinese population. When stratified by age, both old and young patients showed a decrease in HCC risk. When stratified by region, we detected the same association in Chinese in southern China. Similarly when stratified by environment, we observed the same association in Chinese in inland areas; however, no statistically significant association was observed in those in coastal areas.
Conclusion: This meta-analysis suggested that KIF1B (rs17401966) polymorphism could decrease HCC risk in Chinese and in overall population, but not in non-Chinese. This association remained significant in Chinese in southern China and inland areas, but not in those in northern and central China and coastal areas. Further large-scale multicenter studies are warranted to confirm these findings.
Keywords: KIF1B, rs17401966, hepatocellular carcinoma, polymorphism
Background
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor and the second leading cause of cancer-related deaths in the world.1 The onset of HCC is relatively insidious; in most cases, HCC is diagnosed at advanced stages and is difficult to treat. Presently, surgical resection-based comprehensive treatment is the main treatment for HCC, but with less success rate and high rates of recurrence and metastasis.2 Therefore, improving the early diagnosis is particularly important in the prevention and treatment of HCC. Determining the association between KIF1B (rs17401966) polymorphism and HCC risk provides a promising approach to achieve this goal.
KIF1B is a member of the kinesin superfamily and belongs to N-kinesin, encoding two alternatively spliced isoforms, KIF1Bα and KIF1Bβ. Both the isoforms have the same 660 amino acid residues in the N terminal domain; the main difference between them is the end of the C binding domain, conferring different axonal cargo specificity.3 KIF1B is located in chromosome region 1p36.22 and is an important molecule for intracellular vesicle trafficking and organelle transporting.4,5 In addition to transport function, KIF1B also plays an important role in tumor suppression by promoting apoptosis.6 Studies have shown that deficiency of 1p36 region is very common in the individuals with early-onset HCC, but the phenomenon is not observed in individuals with chronic liver disease. It can be speculated that the abnormal chromosomal regions may be associated with the risk of HCC.7
Through genome-wide association study (GWAS), Zhang et al8 found a significant association between KIF1B rs17401966 polymorphism and HCC, showing that the polymorphism of the site has a protective effect on HCC. However, a consistent conclusion on the correlation between the gene polymorphism and HCC was not reached, which may be caused by differences in race or ethnicity, as well as the difference in sample size.8–18 Therefore, we carried out a meta-analysis of the whole included case–control studies to make a more accurate estimate of the association.
Methods
Literature searching strategy
A comprehensive literature searching for all relevant studies published before March 25, 2018 was conducted in PubMed, Web of Science and the Cochrane Library, using the following keywords: KIF1B/Kinesin family member 1B/rs17401966 and locus/mutation/variant*/genotype/polymorphism*/SNP and ([liver/hepatic/hepatocellular/hepato-cellular and carcinom*/cancer/neoplasm*/malign*/tumor] or HCC or hepatoma*) and the combinations. The relevant bibliographies of identified studies were examined for additional articles. Abstracts and citations were screened by two researchers independently, and any disagreements were resolved by discussing with a third reviewer. The full text of all the eligible articles was reviewed during a second screening. There were no language limitations during the retrieval procedure.
Selection and exclusion criteria
All eligible studies included in this meta-analysis met the following inclusion criteria: 1) independent case–control studies performed on humans; 2) evaluated the association between KIF1B (rs17401966) polymorphism and HCC risk; 3) genotype frequencies in case and control groups were available for risk estimate; 4) the diagnosis of the cases was based on pathology; 5) control subjects had no cancer and history of radiotherapy or chemotherapy; and 6) genotype frequencies of the subjects in control groups were in accordance with Hardy–Weinberg equilibrium (HWE). We excluded abstracts, case reports, letters, comments, editorials, reviews, meta-analyses and studies lacking sufficient data. Simultaneously, if the researches were duplicated or shared in more than one study, the most recent publications were included.
Data extraction and synthesis
We used endnote bibliographic software (EndNote X6) to construct an electronic library of citations identified in the literature search. Duplicates were found automatically by endnote and deleted manually. All the extracted data were checked and evaluated twice according to the inclusion criteria listed above by two independent investigators. The following data were extracted from each study: first author, year of publication, country, ethnicity, genotyping method, number of cases and controls, genotype distribution of cases and controls and P-value of HWE in controls. Meanwhile, multicenter studies were divided into several separate studies according to the origin. A third reviewer participated if some disagreements emerged, and a final decision was not made until a consensus was reached.
Quality assessment
The methodological quality assessment was performed based on the modified scoring system used for studies on genetic epidemiological issues.19 Points were awarded on the basis of representativeness of cases, source of controls, HWE in controls, genotyping examination and association assessment. Total score ranged from 0 (lowest quality) to 8 (highest quality). A study with a score of ≥6 was classified to be of high quality.
Statistical analysis
All statistical analyses were carried out using STATA version 11.0 (StataCorp LP, College Station, TX, USA) and Review Manager version 5.2.0 (The Cochrane Collaboration, 2012). Chi-square test was applied to calculate P-value of HWE in controls, and P>0.05 was considered to be consistent with HWE.20 The association of KIF1B (rs17401966) polymorphism and HCC susceptibility was estimated by pooled ORs with 95% CIs under five different genetic models including allele model, dominant model, recessive model, homozygous genetic model and heterozygous genetic model. Z test was used to assess the significance of the ORs. Both Q-statistic test and I2 test were applied to assess the between-study heterogeneity in this meta-analysis. If there was significant heterogeneity among included studies (P-value of Q-statistic was <0.1, or I2 value was >75%), ORs with corresponding 95% CIs were calculated using the random effects model; otherwise, the fixed effects model was selected.20,21 The subgroup analysis was conducted based on ethnicity and age (>50 years or ≤50 years). For studies with Chinese population, we also conducted subgroup analysis by region and environment. Sensitivity analyses were performed to assess the stability of the results. Each study involved in this meta-analysis was deleted each time to reflect the influence of the individual data exerted on the pooled OR. We used Begg’s funnel plot and Egger’s test (P<0.05 was considered significant) to evaluate the publication bias.22,23 All statistical tests were two-sided, and P<0.05 indicated statistical significance.
Results
Characteristics of the included studies
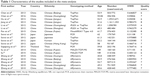
The selection process of eligible studies is presented in Figure 1. A total of 59 relevant articles were preliminarily identified based on our selection strategy. We also identified one article through other sources.18 Thirty-five articles remained after eliminating duplicated literature. Subsequently, 16 obviously irrelevant articles were excluded unquestionably after reviewing their titles and abstracts. Based on the inclusion and exclusion criteria, eight articles were excluded after reviewing the full text. Finally, 11 studies were eventually included in this meta-analysis.8–18 The 11 case–control studies were published between 2010 and 2016. Among them, Zhang et al’s research consisting of five independent studies was divided into five studies.8 Similarly, Li et al’s and Sawai et al’s articles were divided into two and four studies, respectively.13,15 Thus, a total of 19 studies from 11 articles with 8,741 cases and 10,812 controls were included in this meta-analysis. A summary of the characteristics of the 19 studies, including first author, year of publication, country, ethnicity, genotyping method, age of cases, number of cases and controls, P-value of HWE and quality score, is shown in Table 1. Based on quality assessment, all studies were considered to be of high quality (quality scores of these studies were 6–8).
  | Figure 1 Flowchart of studies selection in this meta-analysis. |
Meta-analysis results
The genotype distribution and allele frequencies of KIF1B (rs17401966) polymorphism in cases and controls are listed in Table 2. The main results of our study are shown in Tables 3 and 4.
  | Table 2 KIF1B (rs17401966) polymorphisms genotype distribution and allele frequency in cases and controls |
  | Table 3 Overall meta-analysis results with subgroup conducted by ethnicity and age |
  | Table 4 Subgroup meta-analysis results of Chinese conducted by region and environment |
As shown in Table 3 and Figure 2, the pooled results indicated that the association between KIF1B (rs17401966) polymorphism and the decreased occurrence of HCC was significant in overall population in three genetic models: allele model (OR=0.87, 95% CI=0.78–0.97, P=0.01), dominant model (OR=0.84, 95% CI=0.74–0.94, P=0.003) and heterozygote comparison (OR=0.84, 95% CI=0.76–0.93, P=0.0009). The subgroup analysis stratified by ethnicity showed the same association in Chinese population (allele model: OR=0.84, 95% CI=0.74–0.96, P=0.009; dominant model: OR=0.81, 95% CI=0.71–0.93, P=0.003; homozygous genetic model: OR=0.74, 95% CI=0.56–0.98, P=0.03; heterozygote comparison: OR=0.83, 95% CI=0.74–0.93, P=0.001) (Figure 3), while no genetic model showed significant association in non-Chinese. When stratified by age, we found that both old (allele model: OR=0.86, 95% CI=0.76–0.98, P=0.02; dominant model: OR=0.83, 95% CI=0.73–0.95, P=0.006; heterozygote comparison: OR=0.85, 95% CI=0.76–0.94, P=0.002) and young patients (allele model: OR=0.75, 95% CI=0.59–0.97, P=0.03; dominant model: OR=0.73, 95% CI=0.56–0.96, P=0.03; recessive model: OR=0.64, 95% CI=0.41–0.99, P=0.04; homozygous genetic model: OR=0.57, 95% CI=0.34–0.95, P=0.03; heterozygote comparison: OR=0.77, 95% CI=0.60–0.97, P=0.03) showed a significant association between KIF1B (rs17401966) polymorphism and decreased HCC risk (Figure 4).
For studies with Chinese population, we also conducted subgroup analysis by region and environment. As shown in Table 4, when stratified by region (northern China, central China, southern China), we detected an association of the KIF1B (rs17401966) polymorphism with decreased HCC risk in Chinese in southern China based on heterozygote comparison (OR=0.78, 95% CI=0.63–0.98, P=0.03) (Figure 5). When stratified by environment (inland areas, coastal areas), we observed an association between decreased HCC risk and KIF1B (rs17401966) polymorphism in Chinese in inland areas (allele model: OR=0.76, 95% CI=0.61–0.96, P=0.02; dominant model: OR=0.73, 95% CI=0.58–0.94, P=0.01; homozygous genetic model: OR=0.60, 95% CI=0.36–0.98, P=0.04; heterozygote comparison: OR=0.77, 95% CI=0.63–0.94, P=0.01) (Figure 6); however, no statistically significant association was observed in those in coastal areas.
Sensitivity analyses
As shown in Table 1, all the studies were in line with the balance of HWE in control groups. To evaluate the stability of our results, we performed sensitivity analysis to assess the effect of each individual study on the pooled ORs. After excluding each study sequentially, the corresponding ORs were not substantially changed, suggesting that the results of our meta-analysis were stable and reliable.
Heterogeneity analysis
Heterogeneity among studies was assessed by Q-statistic. Random effects models were applied if P-value of heterogeneity tests was ≤0.1 or I2 was ≥75% (P≤0.1 or I2≥75%), otherwise, fixed effects models were selected (Tables 3 and 4).
Publication bias
Begg’s test, Egger’s test and funnel plot were all used to evaluate the publication bias of the included studies. No significant publication bias was found in Begg’s and Egger’s test (P>0.05). Funnel plot also indicated that publication bias did not exist with no obvious asymmetry that could be observed (Figure 7).
  | Figure 7 Funnel plot assessing evidence of publication bias from 19 studies (heterozygous genetic model, AG vs GG). |
Discussion
GWASs have been shown to be unbiased and effective in exploring disease phenotype-associated single-nucleotide polymorphism (SNP). Currently, a large number of GWASs have been reported, most of which are about cancer.24 Epidemiological and experimental studies have shown that HCC is a complex disease that occurs due to multiple factors, including viral, environmental and genetic factors. With the same environmental background, a small number of people suffer from HCC, whereas others do not, which also shows the importance of genotype. GWASs have found a number of HCC-associated SNPs, such as K1F1B, MICA, HLA-DQA/DQB, SL47W and so on.12,13,25,26 The existence of genetic etiology of HCC is further confirmed. Identification of HCC susceptibility genes and gene-related molecular mechanisms will provide a theoretical basis for the prevention and clinical diagnosis of HCC and treatment of population at high HCC risk. It is expected to achieve early prevention and individualized treatment of HCC and to improve the therapeutic effect of HCC.
Through GWAS, Zhang et al8 found a significant association between KIF1B rs17401966 polymorphism and HCC, showing that the polymorphism of the site has a protective effect on HCC. However, a consistent conclusion on the correlation between the gene polymorphism and HCC was not reached.8–18 Hence, we performed this meta-analysis aiming to illuminate the association between KIF1B (rs17401966) polymorphism and HCC. The pooled results of our study indicated that the association was significant. Subgroup analysis stratified by ethnicity showed the same association in Chinese population, but not in non-Chinese. All the above results were consistent with the results of the meta-analysis of Zhang et al27 and Wang et al.28 However, the number of included papers in their analysis was less than that in our study. When stratified by age, both old and young patients showed decreased HCC risk, which was consistent with the results of Zhang et al’s27 study. When stratified by region (northern China, central China, southern China), we detected an association between KIF1B (rs17401966) polymorphism and decreased HCC risk in Chinese in southern China. When stratified by environment (inland areas, coastal areas), we observed the same association in Chinese in Inland areas; however, no statistically significant association was observed in those in coastal areas. It was the first subgroup analysis on Chinese population stratified by region and environment.
Zhang et al27 also performed subgroup analysis by gender and found that KIF1B rs17401966 polymorphism was significantly associated with HCC in men but not in women. However, the number of papers from which gender data were extracted for their study was only five, and the sample size of women was extremely small. Therefore, we should interpret the results of their study with caution. Zhang et al27 also performed subgroup analysis based on sample sizes and quality scores and found that rs17401966 polymorphism was significantly associated with reduced HCC risk in studies with large sample size and of high quality; however, no significant associations were found in studies with small sample size and of low quality. However, we should realize that small sample sizes and low-quality scores were sources for this heterogeneity, so subgroup analyses stratified by sample sizes and quality scores may not be appropriate.
Nevertheless, some limitations of our meta-analysis should be addressed. First, we could not obtain all the raw data of the patients and hence could not conduct subgroup analysis by sex, hepatitis, liver function and other variables. We also failed to clarify gene–gene and gene–environment interactions in the occurrence and development of HCC. Second, only published studies were included in this meta-analysis; however, some unpublished papers may exist and conform to our inclusion criteria. Therefore, publication bias may have appeared, although no statistical evidence was found. Third, our research is only a comprehensive analysis of existing data. We did not verify the association through basic experiments. Moreover, the included papers were mostly based on Chinese population; only four papers were about non-Chinese. Therefore, data from large-scale multicenter studies based on non-Chinese population are still needed to confirm the association between KIF1B (rs17401966) polymorphism and HCC.
Conclusion
Our meta-analysis indicates that KIF1B (rs17401966) polymorphism could decrease HCC risk in Chinese and in overall population, but not in non-Chinese. This association remained significant in Chinese in southern China and inland areas, but not in those in northern or central China and in coastal areas. Further large-scale multicenter studies are warranted to confirm our findings.
Acknowledgment
This study was funded by National Natural Science Foundation of China (grant numbers 81170454, 30772049 and 30571765).
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Altekruse SF, McGlynn KA, Dickie LA, Kleiner DE. Hepatocellular carcinoma confirmation, treatment, and survival in surveillance, epidemiology, and end results registries, 1992–2008. Hepatology. 2012;55(2):476–482. | ||
Cherng SH, Huang CY, Kuo WW, et al. GABA tea prevents cardiac fibrosis by attenuating TNF-alpha and Fas/FasL-mediated apoptosis in streptozotocin-induced diabetic rats. Food Chem Toxicol. 2014;65:90–96. | ||
Pinhu L, Qin Y, Xiong B, You Y, Li J, Sooranna SR. Overexpression of Fas and FasL is associated with infectious complications and severity of experimental severe acute pancreatitis by promoting apoptosis of lymphocytes. Inflammation. 2014;37(4):1202–1212. | ||
Thurner EM, Krenn-Pilko S, Langsenlehner U, et al. Association of genetic variants in apoptosis genes FAS and FASL with radiation-induced late toxicity after prostate cancer radiotherapy. Strahlenther Onkol. 2014;190(3):304–309. | ||
Rao H, Ma LX, Xu TT, et al. Lipid rafts and Fas/FasL pathway may involve in elaidic acid-induced apoptosis of human umbilical vein endothelial cells. J Agric Food Chem. 2014;62(3):798–807. | ||
He P, Zhou G, Qu D, Zhang B, Wang Y, Li D. HBx inhibits proliferation and induces apoptosis via Fas/FasL upregulation in rat renal tubular epithelial cells. J Nephrol. 2013;26(6):1033–1041. | ||
Zhang H, Zhai Y, Hu Z, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42(9):755–758. | ||
Chen K, Shi W, Xin Z, et al. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci in a Chinese population. PLoS One. 2013;8(10):e77315. | ||
Chen JH, Wang YY, Lv WB, et al. Effects of interactions between environmental factors and KIF1B genetic variants on the risk of hepatocellular carcinoma in a Chinese cohort. World J Gastroenterol. 2016;22(16):4183–4190. | ||
Hu L, Zhai X, Liu J, et al. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology. 2012;55(5):1426–1431. | ||
Jiang DK, Sun J, Cao G, et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45(1):72–75. | ||
Li S, Qian J, Yang Y, et al. GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 2012;8(7):e1002791. | ||
Pan H, Su C, Lin Y, Niu J. [The relationship between the KIF1B (rs17401966) single nucleotide polymorphism and the genetic susceptibility to hepatocellular carcinoma]. Zhonghua Yu Fang Yi Xue Za Zhi. 2015;49(5):419–423. Chinese [with English abstract]. | ||
Sawai H, Nishida N, Mbarek H, et al. No association for Chinese HBV-related hepatocellular carcinoma susceptibility SNP in other East Asian populations. BMC Med Genet. 2012;13:47. | ||
Sopipong W, Tangkijvanich P, Payungporn S, Posuwan N, Poovorawan Y. The KIF1B (rs17401966) single nucleotide polymorphism is not associated with the development of HBV-related hepatocellular carcinoma in Thai patients. Asian Pac J Cancer Prev. 2013;14(5):2865–2869. | ||
Su C, Lin Y, Niu J, Cai L. Association between polymorphisms in tumor suppressor genes and oncogenes and risk of hepatocellular carcinoma: a case-control study in an HCC epidemic area within the Han Chinese population. Med Oncol. 2014;31(12):356. | ||
Su CH. The Association Study Between HBV Infection, Environmental Factors, Polymorphisms and the Risk of Hepatocellular Carcinoma in Xiamen [doctoral thesis]. Fuzhou: Fujian Medical University; 2015. | ||
Niu YM, Du XY, Cai HX, et al. Increased risks between interleukin-10 gene polymorphisms and haplotype and head and neck cancer: a meta-analysis. Sci Rep. 2015;5:17149. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–D1006. | ||
Matsuura K, Sawai H, Ikeo K, et al; Japanese Genome-Wide Association Study Group for Viral Hepatitis. Genome-wide association study identifies TLL1 variant associated with development of hepatocellular carcinoma after eradication of hepatitis C virus infection. Gastroenterology. 2017;152(6):1383–1394. | ||
Lee MH, Huang YH, Chen HY, et al; REVEAL-HCV Cohort Study Group. Human leukocyte antigen variants and risk of hepatocellular carcinoma modified by HCV genotypes: a genome-wide association study. Hepatology. Epub 2017 Sep 16. | ||
Zhang Z. Association between KIF1B rs17401966 polymorphism and hepatocellular carcinoma risk: a meta-analysis involving 17,210 subjects. Tumour Biol. 2014;35(9):9405–9410. | ||
Wang ZC, Gao Q, Shi JY, et al. Genetic polymorphism of the kinesin-like protein KIF1B gene and the risk of hepatocellular carcinoma. PLoS One. 2013;8(4):e62571. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.