Back to Journals » OncoTargets and Therapy » Volume 9
Association between an insertion/deletion polymorphism in IL-1A gene and cancer risk: a meta-analysis
Received 6 September 2015
Accepted for publication 16 November 2015
Published 18 December 2015 Volume 2016:9 Pages 1—6
DOI https://doi.org/10.2147/OTT.S95887
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Daniele Santini
Ling Ma,1 Ning Zhou2
1Department of Stomatology, 2Department of Anesthesiology, No 454 Hospital, PLA, Nanjing, Jiangsu, People’s Republic of China
Purpose: Previous studies have reported the association of an insertion/deletion (Ins/Del) polymorphism (rs3783553) in the 3' untranslated region of interleukin-1A (IL-1A) with the risk of cancer, such as oral squamous cell carcinoma, nasopharyngeal carcinoma, and cervical carcinoma. However, the results are still inconsistent. The present meta-analysis aimed to clarify the association of IL-1A rs3783553 polymorphism with cancer risk.
Methods: All eligible studies were selected from PubMed, Web of Science, and Chinese National Knowledge Infrastructure up to September 2, 2015. Summary odds ratios (ORs) and 95% confidence intervals (CIs) were used to evaluate cancer risk.
Results: A total of ten case–control studies with 4,514 cases and 6,689 controls were included this meta-analysis. We found that IL-1A rs3783553 polymorphism was significantly associated with cancer risk (Ins/Ins + Ins/Del vs Del/Del: OR =0.79, 95% CI =0.67–0.92; Ins/Ins vs Del/Del: OR =0.61, 95% CI =0.47–0.79; Ins/Ins vs Ins/Del + Del/Del: OR =0.67, 95% CI =0.55–0.83; Ins vs Del: OR =0.81, 95% CI =0.72–0.92). In the stratified analyses, significant effects were found among Asian populations (Ins/Ins + Ins/Del vs Del/Del: OR =0.81, 95% CI =0.69–0.95) and cervical carcinoma (Ins/Ins vs Del/Del: OR =0.51, 95% CI =0.34–0.76; Ins/Ins vs Ins/Del + Del/Del: OR =0.52, 95% CI =0.35–0.78).
Conclusion: Our meta-analysis suggests that the IL-1A rs3783553 polymorphism contributes to susceptibility to cancer. However, well-designed studies with larger sample sizes are required to verify the results.
Keywords: IL-1A, polymorphism, cancer, meta-analysis
Introduction
Cancer has been recognized as one of the leading causes of death worldwide. According to the estimation of GLOBOCAN, ~12.7 million new cases and 7.6 million deaths of cancer had occurred in 2008.1 Cancer is a multifactor disease resulting from the combined effect of genetic susceptibility and environmental factors.2–4 The role of inflammation in carcinogenesis is a pivotal issue. Previous studies have demonstrated that inflammation-associated molecules are associated with a majority of cancer types, and these molecules are activated by various elements related to environment and lifestyle.5
Interleukin-1 (IL-1) family consists of the three key proinflammatory cytokines, including IL-1A, IL-1B, and IL-1 receptor antagonist (IL-1RA). Some studies have suggested that IL-1 contributes to tumor proliferation, angiogenesis, tumor invasion, metastases, and patterns of interactions between malignant cells and the host’s immune system.6–9 IL-1A is involved in numerous immune responses and inflammatory process, and has been reported to play an important role in human carcinogenesis.10
The IL-1A gene is located on chromosome 2q14 and has some identified polymorphisms. An insertion/deletion (Ins/Del) polymorphism (rs3783553, TTCA/-) located in the 3′ untranslated regions of IL-1A, has been suggested to regulate the expression levels of IL-1A through disrupting a binding site for miR-122 and miR-378.11 In addition, early epidemiology studies have reported the association between IL-1A rs3783553 polymorphism and the risk of cancer, including cervical carcinoma, oral squamous cell carcinoma, prostate cancer, ovarian cancer, gastric cancer, papillary thyroid carcinoma, hepatocellular carcinoma, and nasopharyngeal carcinoma.11,12–20 However, results from the published studies remain conflicting rather than conclusive. Therefore, the present study authors performed a meta-analysis to clarify the effects of IL-1A rs3783553 polymorphism on cancer risk.
Materials and methods
Publication search
The present study authors systematically searched the PubMed, Web of knowledge, and Chinese National Knowledge Infrastructure databases. The following medical subject headings terms and free words were used: “interleukin-1A” or “IL-1A” and “polymorphism” or “variant” and “tumor” or “cancer” or “carcinoma”. The last search was updated on September 2, 2015. The search was limited to human, and no language restrictions were applied. Additional studies were identified by a hand search of the references of original studies.
Inclusion and exclusion criteria
The included studies conformed to the following criteria: 1) a case–control study, 2) evaluation of the IL-1A rs3783553 polymorphism and cancer susceptibility, and 3) the inclusion of the detailed data that determine genotype distributions. The major exclusion criteria were: 1) a lack of data regarding the IL-1A rs3783553 polymorphism, 2) the inclusion of duplicate data, and 3) case reports or review articles.
Data extraction
Information was carefully extracted from all eligible studies by two independent investigators (Ling Ma and Ning Zhou). The following data were collected from each study: the first author’s name, year of publication, country of origin, ethnicity, genotyping method, P-values from the Hardy–Weinberg equilibrium test for the control group, and genotype and allele frequencies among the cases and controls. All disagreements were discussed and resolved with consensus.
Statistical analysis
Hardy–Weinberg equilibrium among controls for each study was assessed using Pearson’s χ2 test. The strength of association between IL-1A rs3783553 polymorphism and the risk of cancer was estimated for each study by crude odds ratio (OR) and corresponding 95% confidence interval (CI). Four models were conducted: dominant model (Ins/Ins + Ins/Del vs Del /Del), heterozygote comparison (Ins/Del vs Del/Del), homozygote comparison (Ins/Ins vs Del/Del), recessive model (Ins/Ins vs Ins/Del + Del/Del), and allele model (Ins vs Del). Summary ORs and corresponding 95% CIs were estimated by the fixed-effects model or the random-effects model. The χ2-test-based Q statistic test was performed to assess between-study heterogeneity. The effect of heterogeneity was quantified according to the I2 value. When a significant Q test (P<0.05) or I2>50% indicated heterogeneity across studies, the random-effects model was used. Otherwise, the fixed-effects model was applied. Stratification analyses on cancer type and ethnicity were performed. Sensitivity analysis was tested by sequentially omitting one individual study at a time. Finally, the Begg’s funnel plot and Egger’s test were adopted to assess potential publication bias. All statistical analyses were implemented by the STATA Software (version 9.0, Stata Corp., College Station, TX, USA).
Results
Eligible studies
The study selection procedure is shown in Figure 1. A total of 77 articles were identified that were relevant to the search terms. After screening the titles and abstracts, 64 articles were excluded for being irrelevant to IL-1A polymorphisms and cancers risk. After reading the full texts of the 13 articles, three articles were excluded for being irrelevant to the investigated polymorphism. Finally, ten studies were left for data extraction. The main characteristics of the eligible studies are presented in Tables 1 and 2. A total of 4,514 cancer cases and 6,689 controls were included for data synthesis. In addition, nine studies were conducted in Asian populations and only one in European population.
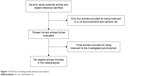  | Figure 1 Flowchart showing study selection procedure. |
  | Table 1 General characteristics of studies included in the meta-analysis |
Quantitative synthesis
As shown in Table 3 and Figure 2, meta-analysis of the total studies suggested that there was a significant association between the IL-1A rs3783553 polymorphism and cancer risk (Ins/Ins + Ins/Del vs Del/Del: OR =0.79, 95% CI =0.67–0.92; Ins/Ins vs Del/Del: OR =0.61, 95% CI =0.47–0.79; Ins/Ins vs Ins/Del + Del/Del: OR =0.67, 95% CI =0.55–0.83; Ins vs Del: OR =0.81, 95% CI =0.72–0.92). The subgroup analysis according to ethnicity suggested that IL-1A rs3783553 polymorphism was significantly associated with cancer risk in Asian populations (Ins/Ins + Ins/Del vs Del/Del: OR =0.81, 95% CI =0.69–0.95). Subgroup analysis by cancer type showed a significant association between IL-1A rs3783553 polymorphism and cervical carcinoma (Ins/Ins vs Del/Del: OR =0.51, 95% CI =0.34–0.76; Ins/Ins vs Ins/Del + Del/Del: OR =0.52, 95% CI =0.35–0.78).
Sensitivity analysis and publication bias
After sequentially excluding each case–control study, statistically similar results were obtained (Figure 3). This suggests that the data of our meta-analysis are relatively stable and credible. In addition, Begg’s funnel plot and Egger’s test were performed to assess the publication bias of our meta-analysis. As shown in Figure 4, the shape of the funnel plot appeared to be symmetrical. The Egger’s test also showed no publication bias.
  | Figure 3 Sensitivity analysis of the IL-1A rs3783553 polymorphism and cancer risk under dominant model. |
  | Figure 4 Begg’s funnel plot of the IL-1A rs3783553 polymorphism and cancer risk under dominant model. |
Discussion
To reveal a small effect of the polymorphisms on cancer risk, a single study might have low statistical power to detect a true association, particularly for studies with small sample size. Meta-analysis is considered as a powerful tool to summarize inconclusive results from different studies and produce a single estimate with enhanced precision. In the current meta-analysis, we found that the IL-1A rs3783553 polymorphism was statistically significantly associated with decreased risk of cancer under dominant model, homozygote comparison, recessive model, and allele model. In addition, stratification analyses by ethnicity and tumor type showed significant association in Asian populations and cervical carcinoma. Although previous functional experiment has shown that IL-1A rs3783553 polymorphism can regulate the expression levels of IL-1A through disrupting a binding site for miR-122 and miR-378 in hepatocellular carcinoma. However, whether the SNP has identical function in different cancer types needs to be further investigated.11
To the best of our knowledge, this was the first meta-analysis providing comprehensive insights into the effects of IL-1A rs3783553 polymorphism on cancer risk. However, some limitations of this meta-analysis study should be considered. First, due to lack of original data, we could not evaluate the potential interactions of gene–gene and gene–environment. Second, the number of the eligible studies was small, which may result in potential publication bias, in spite of no significant publication bias in our meta-analysis. Finally, this meta-analysis included data mainly from Asian populations, so that, the results are applicable to specific ethnic groups.
Conclusion
The current meta-analysis results suggest that the IL-1A rs3783553 polymorphism is associated with susceptibility to cancer. However, larger well-designed studies are warranted to validate these findings.
Disclosure
The authors report no conflicts of interest in this work.
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. | ||
Dai ZM, Fei YL, Zhang WG, et al. Association of vitamin D receptor Cdx-2 polymorphism with cancer risk: a meta-analysis. Medicine (Baltimore). 2015;94(33):e1370. | ||
Liu H, Jia J, Mao X, Lin Z. Association of CYP1A1 and GSTM1 polymorphisms with oral cancer susceptibility: a meta-analysis. Medicine (Baltimore). 2015;94(27):e895. | ||
Peng Q, Li S, Lao X, Chen Z, Li R, Qin X. Association between XPD Lys751Gln and Asp312Asn polymorphisms and hepatocellular carcinoma risk: a systematic review and meta-analysis. Medicine (Baltimore). 2014;93(29):e330. | ||
Sethi G, Shanmugam MK, Ramachandran L, Kumar AP, Tergaonkar V. Multifaceted link between cancer and inflammation. Biosci Rep. 2012;32(1):1–15. | ||
Pantschenko AG, Pushkar I, Anderson KH, et al. The interleukin-1 family of cytokines and receptors in human breast cancer: implications for tumor progression. Int J Oncol. 2003;23(2):269–284. | ||
Elaraj DM, Weinreich DM, Varghese S, et al. The role of interleukin 1 in growth and metastasis of human cancer xenografts. Clin Cancer Res. 2006;12(4):1088–1096. | ||
Gemma A, Takenaka K, Hosoya Y, et al. Altered expression of several genes in highly metastatic subpopulations of a human pulmonary adenocarcinoma cell line. Eur J Cancer. 2001;37(12):1554–1561. | ||
Liss C, Fekete MJ, Hasina R, Lam CD, Lingen MW. Paracrine angiogenic loop between head-and-neck squamous-cell carcinomas and macrophages. Int J Cancer. 2001;93(6):781–785. | ||
Kasza A. IL-1 and EGF regulate expression of genes important in inflammation and cancer. Cytokine. 2013;62(1):22–33. | ||
Gao Y, He Y, Ding J, et al. An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1alpha 3′ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis. 2009;30(12):2064–2069. | ||
Pu Y, Zhang Z, Zhou B, et al. Association of an insertion/deletion polymorphism in IL1A 3′-UTR with risk for cervical carcinoma in Chinese Han Women. Hum Immunol. 2014;75(8):740–744. | ||
Huang J, Ni S, Li D, He Y. An insertion/deletion polymorphism at miRNA-122 binding site in the IL1A is associated with a reduced risk of cervical squamous cell carcinoma. Genet Test Mol Biomarkers. 2015;19(6):331–334. | ||
Zhang Y, Sturgis EM, Sun Y, et al. A functional variant at miRNA-122 binding site in IL-1a 3′ UTR predicts risk and HPV-positive tumours of oropharyngeal cancer. Eur J Cancer. 2015;51(11):1415–1423. | ||
Liao H, Zhang L, Cheng P, et al. Insertion/deletion polymorphism of IL1A 3′-UTR associated with the susceptibility of prostate cancer. Sichuan Da Xue Xue Bao Yi Xue Ban. 2014;45(6):956–959. | ||
Zhang Z, Zhou B, Gao Q, et al. A polymorphism at miRNA-122-binding site in the IL-1a 3′UTR is associated with risk of epithelial ovarian cancer. Fam Cancer. 2014;13(4):595–601. | ||
Zeng XF, Li J, Li SB. A functional polymorphism in IL-1A gene is associated with a reduced risk of gastric cancer. Tumour Biol. 2014;35(1):265–268. | ||
Gao L, Zhu X, Li Z, et al. Association between a functional insertion/deletion polymorphism in IL1A gene and risk of papillary thyroid carcinoma. Tumour Biol. 2014;35(4):3861–3865. | ||
Du Y, Han X, Pu R, Xie J, Zhang Y, Cao G. Association of miRNA-122-binding site polymorphism at the interleukin-1 α gene and its interaction with hepatitis B virus mutations with hepatocellular carcinoma risk. Front Med. 2014;8(2):217–226. | ||
Yang ZH, Dai Q, Zhong L, Zhang X, Guo QX, Li SN. Association of IL-1 polymorphisms and IL-1 serum levels with susceptibility to nasopharyngeal carcinoma. Mol Carcinog. 2011;50(3):208–214. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



