Back to Journals » Infection and Drug Resistance » Volume 15
Assessing Colistin Resistance by Phenotypic and Molecular Methods in Carbapenem-resistant Enterobacterales in a Tertiary Care Hospital in Pakistan
Authors Furqan W , Ali S, Usman J, Hanif F, Naeem A, Nasrullah A , Tayyab N
Received 13 June 2022
Accepted for publication 25 August 2022
Published 10 October 2022 Volume 2022:15 Pages 5899—5904
DOI https://doi.org/10.2147/IDR.S376490
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Warda Furqan, Sakhawat Ali, Javaid Usman, Faisal Hanif, Afnan Naeem, Amnah Nasrullah, Nadia Tayyab
Department of Microbiology Army Medical College (National University of Medical Sciences), Rawalpindi, Pakistan
Correspondence: Warda Furqan, House No. 4, Street No.18, Sector H DHA2, Islamabad, Pakistan, Tel +92-336-5141818, Email [email protected]
Introduction: Members of Enterobacterales are very common pathogens, which continue to show resistance to many antibiotics. Carbapenem performed well for some time. Colistin was the final hope for the carbapenem-resistant Enterobacterales, but resistance against it has virtually tied the clinician’s hands, especially when it comes to treating critically ill patients.
Purpose: Detection of colistin resistance by the agar method as well as by the polymerase chain reaction (mobilized colistin resistance-1 gene) in carbapenem-resistant Enterobacterales.
Materials and Methods: A cross-sectional study from Dec 2019 to Dec 2020 was conducted at the Department of Microbiology, Army Medical College, National University of Medical Sciences Rawalpindi Pakistan. Antimicrobial susceptibility of Enterobacterales was determined according to the Kirby–Bauer disc diffusion method except for colistin. Colistin agar was used, in concentrations of 2 μg/mL and 4 μg/mL. Results were interpreted according to Clinical and Laboratory Standards Institute guidelines 2020. Mobilized colistin-resistant-1 gene in the carbapenem resistant Enterobacterales was detected by performing real-time polymerase chain reaction assay.
Results: Among the 172 carbapenem-resistant Enterobacterales 18 isolates were resistant using the colistin agar test. Whereas by molecular method colistin resistance was detected among 10 isolates that carried mobilized colistin resistance 1 gene, making the frequency of the MCR-1 gene 5.81%. Seventy percent of isolates were from paired blood samples. Eight patients, from whom the colistin resistant gene was isolated expired.
Conclusion: Colistin resistance is a very serious issue and should not be missed in a clinical microbiology laboratory. The phenotypic agar test method is an excellent option for routine use, as it combines ease of performance with affordable cost. However, molecular methods are essential for the detection of mobilized colistin resistance gene (1– 9) for epidemiological purposes.
Keywords: carbapenem-resistant Enterobacterales, colistin agar test, Enterobacterales, mobilized colistin resistant-1 gene, phenotypic method, real time polymerase chain reaction
Introduction
The rate at which resistance against carbapenems is occurring poses a serious threat to the health-care systems globally. The situation is even more serious in developing countries like Pakistan which has a population of over 200 million. The facility of functioning clinical microbiology laboratories is available in a few major cities.1 Clinicians, due to this problem, are forced to treat their patients on an empiric basis. The situation gets aggravated when antibiotics are freely available over the counter.2
Carbapenems, the broad-spectrum bactericidal ß-lactam group of antibiotics were available just in time to rescue when several gram-negative rods were developing multidrug resistance. However, these organisms did not take very long to render carbapenems ineffective.3
Resistance against carbapenems is due to four major mechanisms which include the production of low affinity penicillin-binding protein (PBPs) targets, reduced outer membrane permeability, efflux mechanism, and production of ß-lactamase.4–6 Carbapenem resistance in Enterobacterales is a major issue, particularly in resource-limited countries.
Colistin is an old polycationic, bactericidal antibiotic, targeting the lipid A portion of lipopolysaccharides, moving its cationic charges and leading to cell wall lysis and bacterial death.7 It was the next and almost last hope for treating CRE.8 Testing bacteria against colistin in a microbiology laboratory has some peculiarities eg in 2019 CLSI recommended broth microdilution for it, a laborious process and not done at many laboratories.9,10 Alternatively, it was tested with a colistin disc of 10 µg with other antibiotics by the Kirby–Bauer disc diffusion method. The recommendations later were shifted to the colistin agar test method in 2020.11 Previously resistance against colistin was known to be chromosomally mediated which caused the change in membrane permeability and efflux pump for antibiotics.
Chromosomes were considered the main reason for colistin resistance till the reporting of the plasmid mediated MCR-1 gene in 2015.12 The MCR-1 gene was detected in clinical isolates in China its prevalence was 4.8% according to a longitudinal study.13 Whereas in Pakistan 10 MCR-1 genes were detected out of 52 colistin resistant isolates.14 Colistin resistance indeed foretold a catastrophe.
Keeping all these points under consideration we set out to detect colistin resistance among the clinically isolated CRE, both by CLSI recommended agar method as well as the detection of the MCR-1 gene by real-time PCR.
Materials and Methods
This cross-sectional study was conducted from Dec 2019 to Dec 2020 at the Department of Microbiology, Army Medical College, National University of Medical Sciences Rawalpindi Pakistan. One hundred and seventy-two clinical samples of CRE isolated from pus, blood, body fluids respiratory tract aspirates specimens, and tissue received for culture and sensitivity in the Laboratory of Microbiology, Army Medical College affiliated with the National University of Medical College were processed according to standard microbiological guidelines.
The study was reviewed and approved by the Research and Ethics Review Committee of Army Medical College. A written permission letter was also obtained from the committee. The purpose and procedures of the study were explained. A written consent was taken from all participants. The confidentiality of all study participants was maintained via the study participant secret code. For participants who cannot read and write, the information sheets were read to them. This research was carried out in accordance with the Declaration of Helsinki.
Specimens were inoculated on blood agar, MacConkey agar and chocolate agar according to the requirement of specimen. The agar plates were incubated for 24 to 48 h at 35°C to 37°C in ambient air. Initial identification of bacterial growth was done by colony morphology and Gram staining. Further for gram-negative rods oxidase, catalase, and motility tests were performed. Biochemical identification of gram-negative rod was done by Analytical Profile Index for Enterobacteriaceae, bioMérieux (API20E). The modified Kirby–Bauer disc diffusion method was used for antimicrobial susceptibility testing. Antimicrobial discs of different concentrations for Enterobacteriaceae were used as per Clinical and Laboratory Standard Guidelines (CLSI) guidelines 2020.
Colistin susceptibility was phenotypically assessed by colistin agar test. A stock solution of colistin sulphate was prepared using 5.2 mg colistin sulphate powder was added to 100 mL of distilled water to get final concentration of 40 µg/mL. This solution was divided in 5 mL aliquots. For 2 µg/mL one colistin stock solution bottle was added in 95 mL sterilized agar. For 4 µg/mL plates add two 5 mL aliquots in 90 mL of sterilized agar. Both the plates were inoculated with the isolates. Any growth was considered as colistin resistant and no growth was considered as colistin-sensitive interpretations were as CLSI 2020 guidelines.
The isolated CRE were subjected to real-time polymerase reaction for the detection of MCR-1 gene. The primers were designed using Primer3Plus software and compatibility was checked by NCBI-blast. A forward (5’-AAGGCGTGATGGATAAGCTG-3’) and a reverse primer (5’-TGCAGCCACTGGATACTTTG-3’) of the aforementioned sequence were used. Thermo Scientific GeneJET Genomic DNA purification kit was used for manual extraction of DNA. PCR was done using Smart cycler by Cepheid. Denaturation was done at 94°C for 10 min and later for 30 seconds. Primer annealing and extension and was done at 68°C for 45 seconds. Final extension step was carried out at 72°C for 5 min. The samples were considered negative if no fluorescent signals were detected.
Data was analyzed using Statistical Package for Social Sciences (SPSS) version 22. P<0.001 was considered significant. Frequency and percentages were calculated for qualitative variables. Quantitative variables were presented as mean and standard deviation (SD).
Results
During the one year study 172 clinical isolates were analyzed. Out of these total samples, the highest number of the CRE were isolated from the blood specimens (n=120) followed by lower respiratory tract specimens (n=33). The remaining CRE were isolated from pus, body fluids, and tissue as shown in Table 1.
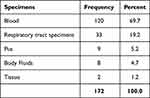 |
Table 1 Frequency and Percentage of CRE Isolated from Specimen for Bacterial Culture |
Among CRE Klebsiella pneumoniae and Escherichia coli were the two most common species accounting for 64.5% and 17.4% respectively. Other carbapenem resistant Enterobacteriaceae included Enterobacter aerogenes, Klebsiella oxytoca and Citrobacter freundii as shown in Figure 1.
 |
Figure 1 Carbapenem Resistant Enterobacterales isolated from specimens and frequency of MCR-1 gene in isolates. |
All the CRE were isolated from hospitalized patients with an average duration of 8.5 days of stay in a critical care unit. Among 172 clinical specimens 68 were received from the neonatal intensive care unit (ICU) and the remaining 104 were from adult intensive care units.
Among the CRE 18 isolates were resistant to colistin by agar test using 2 to 4 µg/mL concentrations whereas 10 CRE isolates were found to possess the MCR-1 gene by the real time PCR. As shown in the Table 2.
 |
Table 2 Details of Ten Patients Infected with MCR-1 Gene Carrying CRE |
Table 2 shows that 07 out of 10 (70%) organisms carrying the MCR-1 gene were isolated from blood specimens and 03 (30%) were from bronchoalveolar lavage (BAL) specimens. Analytical test was applied to calculate the significance between different variables. The days of hospital stay to colistin resistance had a P value of 0.001. Ten samples from which the MCR-1 gene was detected, contained from 7 patients were admitted in the adult intensive care units who were on mechanical ventilation for more than 7 days, and three from the neonatal ICU. Out of 10 only two patients survived in which the MCR-1 gene was detected.
Discussion
Indiscriminate use of antibiotics over the years gave origin to multidrug resistant organisms and extensively drug resistant organisms which are a major threat to health care system worldwide.15 With development of newer antibiotics and combination therapies, bacteria have also modified themselves and have developed various resistance mechanisms. This leaves a physician with limited therapeutic options, especially in developing countries where infectious diseases are rampant and many times microorganisms are present in their most resistant forms. This situation has becomes more alarming for patients in critical care units.16
In our study the carbapenem resistant Klebsiella pneumoniae accounting for highest proportion (82.05%) goes in parallel with studies conducted in Nanjing district of China, Taiwan and Henan China in which the proportion of carbapenem resistant Klebsiella pneumoniae was 79.4%,17 76.3%18 and 52.9%19 respectively.
Chronically ill patients require medical instruments like ventilators and devices like urinary catheters, IV catheters. These devices usually get colonized by resistant bacteria. Patients with repeated hospital admissions also require broad spectrum antibiotics and are more prone to be infected with resistant bacteria.20,21 As in our study, all patients were admitted to the hospital for more than 4 days and 100 patients were on ventilators.Also proven by our study results and according to statistical analysis prolonged hospital stay to colistin resistance was more significant.
In our study PCR results showed that among one hundred and seventy two CRE isolated, MCR-1 was detected in ten isolates (5.8%) only. The percentage of MCR-1 gene in our study is lower as compared to another study conducted in Pakistan in which percentage was 11.4%.22 Our results are comparable to studies conducted in Iran, Thailand and China in where percentage of MCR-1 gene was 1.7%, 4.4% and 6.8% respectively.23–25 Not very long ago no clear guidelines were available for the disc method and the CLSI recommended broth dilution only, a cumbersome and relatively expensive procedure, requiring trained manpower. Lately the use of colistin agar has been recommended for the detection of colistin resistance.
Using phenotypic and molecular method we found out a difference of 8 isolates as 18 were picked by the agar and in 10 isolates MCR-1 gene was detected by PCR. The most probable explanation is that these eight isolates colistin resistant due to MCR 2–9 genes. We could not go for a multiplex PCR for their detection, due to the limited resources. And keeping in view the epidemiology of MCR genes globally MCR-1 prevalence was the highest.26,27
In our study out of the 10 isolates in which MCR-1 gene was detected, 6 were Klebsiella pneumoniae and 4 isolates were Escherichia coli. Thus organism isolated in our study corresponds to a study in which 4 pathogens carrying the MCR-1 gene were detected, two were Klebsiella pneumoniae and two isolates were Escherichia coli.28
All MCR-1 gene were isolated from blood samples which go in parallel with a meta-analysis conducted in China from 2014 to 2019 which showed in two studies one from 2015 and another from 2019 that all MCR-1 gene carrying organisms were isolated from the blood specimens.29
Our study indicates that colistin being used most frequently for critically ill patients has raised resistance against drug of last resort in Pakistan. Outbreaks can occur in wards if appropriate steps are not taken towards implementation of antimicrobial stewardship. Targeted antimicrobial therapy according to culture and sensitivity along with clinical correlation is the need of hour for control of these resistant pathogens. As resistance due to MCR-1 gene is transferable through plasmid so infection prevention and control (IPC) remains one of the major elements in controlling antimicrobial resistance, by breaking the cycle of transmission of resistant organisms. With a population of over 200 million we have limited functioning clinical microbiology laboratories, and where available the antibiotic sensitivity is carried out on the conventional Kirby-Bauer method. The molecular detection is extremely limited.30
Healthcare facilities face a lack of infection control practices as well as infrastructure for isolation. These all lead to a rapid rise in Health-care Associated Infections (HCAIs) in Pakistan. According to a study, the prevalence of HCAIs among neonates was 23.8% and in adult, intensive care units were 33.3%.31
There is a dire need to conduct multicenter studies in our healthcare centers for the detection of the MCR-1 gene along with other variants 2, 3, 4, 6, 7, 8 and 9 detected by multiplex PCR.
Conclusion
Colistin resistance is a very serious issue and should not be missed in a clinical microbiology laboratory. The phenotypic agar test method is an excellent option for routine use, as it combines ease of performance with affordable cost. However, the molecular methods are essential for the detection of mobilized colistin resistance gene (1–9) for epidemiological purposes.As we gain access to more resources, we will look for the non -MCR-1 genes confirming colistin resistance. Antimicrobial stewardship should be strictly implemented to avoid further development of resistance.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lutgring JD. Carbapenem-resistant Enterobacteriaceae: an emerging bacterial threat. Semin Diagn Pathol. 2019;36(3):182–186. doi:10.1053/j.semdp.2019.04.011
2. Aslam A, Gajdács M, Zin CS, et al. Evidence of the practice of self-medication with antibiotics among the lay public in low- and middle-income countries: a scoping review. Antibiotics. 2020;9(9):597. doi:10.3390/antibiotics9090597
3. Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin Infect Dis. 2019;69(Supplement_7):S521–S528. doi:10.1093/cid/ciz824
4. Li Y, Sun QL, Shen Y, et al. Rapid increase in prevalence of carbapenem-resistant enterobacteriaceae (CRE) and emergence of colistin resistance gene mcr-1 in CRE in a hospital in Henan, China. J Clin Microbiol. 2018;56(4). doi:10.1128/JCM.01932-17
5. Richter S, Virulence DM. Screening for carbapenem-resistant Enterobacteriaceae: who, when, and how? Taylor Fr. 2016;8(4):417–426. doi:10.1080/21505594.2016.1255381
6. van Duin D. Carbapenem-resistant Enterobacteriaceae: what we know and what we need to know. Virulence. 2017;8(4):379–382. doi:10.1080/21505594.2017.1306621
7. Hancock REW, Chapple DS. Peptide antibiotics. Antimicrob Agents Chemother. 1999;43(6):1317–1323. doi:10.1128/AAC.43.6.1317
8. Giacobbe DR, Saffioti C, Losito AR, et al. Use of colistin in adult patients: a cross-sectional study. J Glob Antimicrob Resist. 2020;20:43–49. doi:10.1016/j.jgar.2019.06.009
9. Ezadi F, Ardebili A, Mirnejad R. Antimicrobial susceptibility testing for polymyxins: challenges, issues, and recommendations. J Clin Microbiol. 2019;57(4). doi:10.1128/JCM.01390-18
10. Wanyne P. Clinical and Laboratory Standard Institute. Vol. M100.
11. Wayne P. Clinical Laboratory Standard Institute. Vol. M100. CLSI; 2020.
12. Liu -Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
13. Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
14. Hameed F, Khan M; HB- … C& H. Detection of MCR-1 gene in multiple drug resistant Escherichia coli and Klebsiella pneumoniae in human clinical samples from Peshawar, Pakistan; 2021. ingentaconnect.com. Available from: https://www.ingentaconnect.com/content/ben/cchts/2021/00000024/00000005/art00011.
15. Naylor NR, Pouwels KB, Hope R, et al. The health and cost burden of antibiotic resistant and susceptible Escherichia coli bacteraemia in the English hospital setting: a national retrospective cohort study. PLoS One. 2019;14(9):e0221944. doi:10.1371/journal.pone.0221944
16. Peters L, Olson L, Khu DTK, et al. Multiple antibiotic resistance as a risk factor for mortality and prolonged hospital stay: a cohort study among neonatal intensive care patients with hospital-acquired infections caused by gram-negative bacteria in Vietnam. PLoS One. 2019;14(5):e0215666. doi:10.1371/journal.pone.0215666
17. Zhou H, Zhang K, Chen W, et al. Epidemiological characteristics of carbapenem-resistant Enterobacteriaceae collected from 17 hospitals in Nanjing district of China. Antimicrob Resist Infect Control. 2020;9(1). doi:10.1186/S13756-019-0674-4
18. Chen C-W, Tang H-J, Chen -C-C, et al. The microbiological characteristics of carbapenem-resistant Enterobacteriaceae carrying the mcr-1 gene. J Clin Med. 2019;8:261. doi:10.3390/jcm8020261
19. Munoz-Price L, Poirel L; RB-TL Infectious. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases; 2013. Elsevier. Available from: https://www.sciencedirect.com/science/article/pii/S1473309913701907.
20. Hayden M, Lin M, Lolans K; SW-CI. Prevention of colonization and infection by Klebsiella pneumoniae carbapenemase–producing Enterobacteriaceae in long-term acute-care hospitals; 2015. academic.oup.com. Available from: https://academic.oup.com/cid/article-abstract/60/8/1153/2462777.
21. Thaden J, Lewis S, Hazen K; KH-IC&. Rising rates of carbapenem-resistant enterobacteriaceae in community hospitals: a mixed-methods review of epidemiology and microbiology practices in a network of; 2014. cambridge.org. Available from: https://www.cambridge.org/core/journals/infection-control-and-hospital-epidemiology/article/rising-rates-of-carbapenemresistant-enterobacteriaceae-in-community-hospitals-a-mixedmethods-review-of-epidemiology-and-microbiology-practices-in-a-network-of-community-hospitals-in-the-southeastern-united-states/1E94FDE8AE9F631E92A329D7FACE591E.
22. Bilal H, Tu R, Khan M, et al. Discovery of mcr-1 harboring Incl2 plasmids from clinical isolates of multiclonal E. coli prevalent in Pakistan; 2020. Available from: https://scholar.archive.org/work/r2khneqzfzfkxhwkh3cw3lm4hi/access/wayback/https://assets.researchsquare.com/files/rs-74913/v1/9822f58b-9654-4817-9b86-43f53d9d1301.pdf.
23. Moosavian M; Resistance NE-I and Drug. The first report of emerging mobilized colistin-resistance (mcr) genes and ERIC-PCR typing in Escherichia coli and Klebsiella pneumoniae clinical isolates in; 2019. ncbi.nlm.nih.gov. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/pmc6500874/.
24. Zhong L, Phan H, Shen C; KV-CI. High rates of human fecal carriage of mcr-1–positive multidrug-resistant Enterobacteriaceae emerge in china in association with successful plasmid families; 2018. academic.oup.com. Available from: https://academic.oup.com/cid/article-abstract/66/5/676/4430429.
25. Eiamphungporn W, Yainoy S; CJ-J of Global. Prevalence of the colistin resistance gene mcr-1 in colistin-resistant Escherichia coli and Klebsiella pneumoniae isolated from humans in Thailand; 2018. Elsevier. Available from: https://www.sciencedirect.com/science/article/pii/S2213716518301176.
26. Ling Z, Yin W, Shen Z, Wang Y, Shen J, Walsh TR. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother. 2020;75(11):3087–3095. doi:10.1093/jac/dkaa205
27. Wise MG, Estabrook MA, Sahm DF, Stone GG, Kazmierczak KM. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014–2016 as part of the INFORM global surveillance program. PLoS One. 2018;13(4):e0195281. doi:10.1371/journal.pone.0195281
28. Du H, Chen L, Tang YW, Kreiswirth BN. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis. 2016;16(3):287–288. doi:10.1016/S1473-3099(16)00056-6
29. Huang H, Dong N, Shu L, et al. Colistin-resistance gene mcr in clinical carbapenem-resistant Enterobacteriaceae strains in China, 2014–2019. Emerg Microbes Infect. 2020;9(1):237–245. doi:10.1080/22221751.2020.1717380
30. Jiang Y, Zhang Y, Lu J, et al. Clinical relevance and plasmid dynamics of mcr-1-positive Escherichia coli in China: a multicentre case-control and molecular epidemiological study. Lancet Microbe. 2020;1(1):e24–e33. doi:10.1016/S2666-5247(20)30001-X
31. Saleem Z, Hassali MA, Godman B, Hashmi FK, Saleem F. A multicenter point prevalence survey of healthcare–associated infections in Pakistan: findings and implications. Elsevier. Available from: https://www.sciencedirect.com/science/article/pii/S0196655318309556.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
