Back to Journals » OncoTargets and Therapy » Volume 13
ASH2L-Promoted HOXC8 Gene Expression Plays a Role in Mixed Lineage Leukemia-Rearranged Acute Leukemia
Authors Wu YJ, Li L, Liu L, Zhao S, Qiu H, Wang H
Received 2 July 2019
Accepted for publication 3 December 2019
Published 14 January 2020 Volume 2020:13 Pages 381—387
DOI https://doi.org/10.2147/OTT.S221643
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Yu-Jie Wu, 1 Li-xia Li, 2 Lu Liu, 1 Si-shu Zhao, 1 Hai-rong Qiu, 1 Hui Wang 1
1Department of Hematology, The First Affiliated Hospital of Nanjing Medical University (Jiangsu Provincial People Hospital), Nanjing 210029, Jiangsu Province, People’s Republic of China; 2Department of Laboratory Medicine Center, The Second Affiliated Hospital of Nanjing Medical University (Jiangsu Provincial People Hospital), Nanjing 210011, Jiangsu Province, People’s Republic of China
Correspondence: Yu-Jie Wu Email [email protected]
Background: Mixed lineage leukemia (MLL) fusion protein alone exhibits poor histone lysine methyltransferase (HKMT) activity in catalyzing histone H3 Lys4 trimethylation (H3K4me3) in MLL-rearranged acute leukemia.
Methods: To explore the HKMT effect of another regulatory protein within the complex of proteins associated with Set 1 (COMPASS), we analyzed the H3K4me3 modification of the HOXC8 promoter under the action of ASH2L regulation. Small interfering RNA of ASH2L, chromatin immunoprecipitation, real-time-PCR (RT-PCR), and Western blotting were used to detect the expression of specific regions of the HOXC8 promoter, RBBP5, WDR5, MLL, and BRTF in two MLL-rearranged acute leukemia cell lines (RS4:11 and THP-1 cells).
Results: The gene and protein expression levels of HOXC8 were significantly downregulated upon treatment with ASH2L-siRNA (as analyzed by targeting specific regions of the HOXC8 promoter located 0 and 3 kb (– 3.0 kb) upstream of the transcriptional start site in RSH:11 cells; and – 3.0 and – 2.0 kb upstream of the transcriptional start site, and +1.4 kb downstream of the transcriptional start site in THP-1 cells). The expression levels of the BRTF, RBBP5, WDR5, and MLL genes were significantly downregulated from the different transcriptional start sites of the HOXC8 promoter in the RSH:11 cell line (P < 0.05). Furthermore, the BPTF and RBBP5 genes were downregulated from the HOXC8 promoter in the THP-1 cell line (P < 0.05).
Conclusion: Based on these results, we suggest a new concept of histone modification of the ASH2L protein in MLL-rearranged acute leukemia, which cannot carry out methyltransferase activity independently. The protein–protein interactions of ASH2L with other COMPASS members, such as MLL, WDR5, RBBP5, and chromatin remodeling factor BRTF, appear to be essential for its role in the activation of HOXC8 gene transcription.
Keywords: MLL-rearranged acute leukemia, HOXC8 gene, ASH2L, COMPASS complex, H3K4 trimethylation
Introduction
The mixed lineage leukemia (MLL) gene is located in q23 of the long arm of chromosome 11 (11q23), the rearrangement of which is a common chromosomal abnormality in adult and childhood acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML). Patients that harbor this gene rearrangement exhibit unique clinical and biological characteristics. The WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues (2008) individually lists “mixed phenotype leukemia (v; 11q23) with MLL rearrangement” as one of the important subtypes of acute leukemia. This type of leukemia is not usually sensitive to conventional chemotherapy and is associated with a low complete remission rate and a short survival period.1–3
The MLL gene, which encodes an important methyltransferase, is involved in MLL gene rearrangement during the development of leukemia through the regulation of downstream target genes.
The proteins ASH2L, WDR5, RBBP5, and Dpy30 together constitute the MLL1 complex (COMPASS), which exhibits strong enzymatic activity.4 As an important regulatory protein, ASH2L is likely involved in transcriptional activation of the HOXC8 gene in MLL-rearranged acute leukemia.5 To explore the histone lysine methyltransferase (HKMT) effect of another regulatory protein within the complex of proteins associated with Set 1 (COMPASS), we examined the expression of ASH2L and the stability of the HOXC8 gene and analyzed H3K4H3K4 trimethylation (H3K4-me3) modification of the HOXC8 promoter under ASH2L regulation in MLL-rearranged leukemia cells.
Materials and Methods
Cell Lines
Human RS4:11 and THP-1 cell lines were purchased from the Shanghai Cell Bank, of the Chinese Academy of Sciences. The cells were placed in a 5% CO2 incubator at 37°C and the cell density was controlled at 0.5–1×106/mL in RPMI 1640 medium with 10% premium fetal bovine serum.
Fluorescence in-situ Hybridization (FISH)
An MLL two-color fluorescent probe (Vysis Inc., USA) was used in the study. The hybridization signals of red and green fluorescence of cells were detected by excitation of the DAPI/TR filter (Texas Red) and observation under a Leica DRMA2 fluorescence microscope, according to a previous method.6 At least 300 cells were analyzed for each sample. Nuclei with clear boundaries and intact structures that were non-overlapping were counted to calculate the percentage of positive cells. The MLL two-color break-point separation probe diagnostic criteria were that cells with MLL rearrangement showed red-green fusion signals (i.e., a red signal and a green signal), whereas normal cells showed two red-green fusion fluorescence signals.
Quantitative Real-Time PCR (qRT-PCR) Detection of the HOXC8 Gene
For all experiments, three independent samples were detected for the RS4:11 and THP-1 cell lines. The high-purity total RNA rapid extraction kit (General, USA) was used to extract total RNA from the cells. The RNA was then reverse-transcribed into cDNA with the RevertAid First Strand cDNA Synthesis Kit (Fermentas, USA). Quantitative RT-PCR was performed using an SYBR Green RT-PCR kit (Bio-Rad, Hercules, CA, USA) with a quantitative PCR instrument (Bio-Rad). The primers were synthesized by Sangon Biotech (Shanghai, China). The primer sequences for amplification of HOXC8 and the internal reference gene GAPDH are listed in Supplemental Table 1. Relative quantitative analysis was performed for HOXC8 according to the PCR amplification curve, and standard curves for the negative and positive controls were also determined for each experiment.
Small Interfering RNA (siRNA) Transfection
The FAM fluorescently labeled siRNA-ASH2L fragments were chemically synthesized and transfected into cells using Lipofectamine 2000 reagent according to the manufacturer’s instructions. Three sets of primers were used for the ASH2L-siRNA group that specifically interfere with the target gene ASH2L (Supplemental Table 2). Cells were harvested 24 hrs after transfection with siRNA, lysed by the addition of 1 mL of Trizol, and then total RNA was extracted by centrifugation and reverse transcribed into cDNA for qPCR. qPCR was performed in triplicate using SuperReal PreMix Plus (SYBR Green; TIANGEN Biotech) on a CFX connect RT-PCR System (Bio-Rad). The expression levels of HOXC8 and ASH2L cDNAs were normalized to β-actin cDNA by the comparative cycle threshold (Ct) method. The primer sequences for HOXC8 and ASH2L are listed in Supplemental Table 3.
Western Blot Analysis After ASH2L siRNA Treatment
Immunoblot signals were detected with Pierce ECL Western blotting substrate (Thermo Fisher). Cells were lysed in RIPA buffer containing protease inhibitors (Beyotime). The proteins were analyzed by SDS-PAGE and transferred to PVDF membrane. The membrane was exposed to X-ray film after incubation with the indicated ASH2L antibody (Abcam; ab181117) and GAPDH was used as a control.
Chromatin Immunoprecipitation (ChIP)
Before or after transfection for 48 h, cells were fixed in formalin and ChIP was performed against anti-H3K4me3, anti-ASH2L, and IgG antibodies (Abcam). ChIP experiments were performed using a SimpleChIP® Plus Sonication Chromatin IP Kit (Cell Signaling Technology) according to the manufacturer’s instructions. Associated DNA isolated via ChIP was then analyzed by quantitative real-time RT-PCR using IQ SYBR Green Supermix (Bio-Rad, cat no:170-8882AP) and primers to the HOXC8 promoter region, MLL, BPTF, RBBP5, and WDR5, which are listed in Supplemental Table 4a and 4b. The PCR products were electrophoresed and sent to Biosune Inc. (Shanghai) for sequencing using the primers listed in Supplemental Table 4c.
Statistical Methods
For FISH, 200 interphase cells were analyzed for each sample. According to the criterion of the number of positive cells > (x±3s), the positive criterion for MLL-rearrangement was determined to be >8.9%. To analyze gene expression, after PCR, the cycle threshold (Ct value) was recorded. Using GAPDH expression as the internal reference, the expression level of the gene of interest was measured according to the equation 2−ΔCT, where ΔCT = mean value of CT of the gene to be measured ˗ mean value of CT of the internal reference gene. The results were automatically calculated by the fluorescence quantitative PCR software Bio-Rad CFX Manager and were reported in the generated PDF files and the EXCEL raw datasheet. The measured data are represented by x ± s, and the data between the two groups were analyzed by a paired t-test: ** P < 0.01, * P < 0.05.
Results
Detection of Both Cell Types by FISH
In total, 300 interphase cells of the RS4:11 and THP-1 cell types were analyzed. More than 90% of the cells showed a positive signal (one red, one green, and one fusion signal), which confirmed two types of MLL rearrangements in 11q23 (Figure 1).
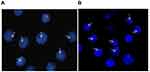 |
Figure 1 FISH detection of the two cell lines of MLL-rearranged acute leukemia. Arrow: positive signal cells (one red, one green, and one fusion signal), (A) THP-1 cells, (B) RS4:11 cells. |
HOXC8 Gene Expression in the Two Cell Lines
HOXC8 gene expression was analyzed in both cell lines by the qPCR method, and the mRNA level in THP-1 cells was found to be 2.7704 (1.0232–6.343), compared with 4.0487 (3.8011–4.2248) in RS4:11 cells. The Ct values of HOXC8 in both cell lines were no more than three times that of the control, indicating a relatively high level of gene expression, and thus the expression of HOXC8 in RS4:11 cells was more stable.
Change in Expression of the ASH2L and HOXC8 Genes Following ASH2L Interference as Determined by Quantitative PCR
After 24 hrs incubation, the green fluorescent proteins carried by the plasmid were observed under a fluorescence microscope to calculate the transfection rate. No green fluorescence was observed in the untransfected group, indicating that the ASH2L gene had been successfully transfected. The results showed that the transfection efficiency of the RS4:11 and THP-1 cells was >90% by inverted fluorescence microscope and >85% by flow cytometry (Figure 2).
Three independent tests confirmed that the expression levels of the ASH2L and HOXC8 genes were downregulated after transfection with ASH2L siRNA. The expression of ASH2L in sample 2 of THP-1 cells and sample 2 of RSH:11 cells was significantly downregulated (P < 0.01); the expression of HOXC8 in sample 2 of THP-1 cells and samples 1, 2, and 3 of RSH:11 cells was also significantly downregulated (P < 0.01), indicating that ASH2L siRNA successfully interfered with the expression of ASH2L in THP-1 and RS4:11 cells. This result also indicated that the expression of HOXC8 was downregulated following ASH2L interference in RSH:11 and THP-1 cells.
qPCR Detection of ASH2L and HOXC8 Gene Expression After siRNA Interference
Three independent qPCR assays showed that the expression of both the ASH2L and HOXC8 genes was downregulated following siRNA interference. The expression of ASH2L was significantly downregulated in sample No. 2 of THP-1 cells and sample No. 2 of RSH:1 cells (P < 0.01); HOXC8 was significantly downregulated (P < 0.01) in sample No. 2 of THP-1 cells and sample No. 1, 2, and 3 of RSH:1 cells (Supplemental Figure 1). The second pair of primers showed the highest efficiency among the three pairs of primers tested so were used in subsequent studies.
Change in ASH2L Protein Expression Following siRNA Interference
Three independent Western blot experiments showed that ASH2L protein expression was downregulated following siRNA interference. The protein expression levels of ASH2L in sample No. 1 and 2 of THP-1 cells and sample No. 2 and 3 of RS4:11 cells were significantly lower than that of the negative control group (P < 0.05), indicating that siRNA successfully interfered with the expression of the target ASH2L protein in THP-1 and RS4:1 cells (Figure 3).
ChIP-qRT-PCR and Sequencing Results Before siRNA Interference
In both RS4:11 and THP-1 cells, the ASH2L protein showed the strongest binding to locus ˗2 kb of the promoter region of HOXC8, although it also showed a degree of enrichment and binding in other regions. In RS4:11 cells, the modification of H3K4me3 was positively correlated with the binding of ASH2L. In THP-1 cells, the modification of H3K4-me3 at locus 1.4 kb of the first intron was elevated abnormally (Supplemental Figure 2). The sequencing results showed that the PCR products correlated with distinct sequences in the HOXC8 promoter (Supplemental Figure 3).
ChIP-qRT-PCR Results After siRNA Interference
After siRNA interference of ASH2L expression, DNA from RS4:11 and THP-1 cells was fixed with formaldehyde and broken into fragments of 200–500 bp by ultrasonic fragmentation. DNA was then extracted by ChIP using a specific negative control antibody (anti-RbIgG) and anti-H3K4-me3. Subsequently, the ˗2, ˗3, and 1.4 kb loci of the promoter region of HOXC8 were detected by qRT-PCR. The results showed that compared with the negative control, modification at locus ˗3 kb of HOXC8 and the transcriptional start region of H3K4-me3 were significantly reduced. In THP-1 cells, modification of loci ˗2 and ˗3 kb of the HOXC8 promoter region and H3K4-me3 of locus 1.4 kb of the first intron were significantly reduced (P < 0.05) (Supplemental Figure 4).
Activity of MLL, BPTF, RBBP5, and WDR5 Was Reduced in the HOXC8 Promoter Region After Treatment with siRNA-ASH2L
After siRNA-ASH2L treatment of RS4:11 cells, decreased activity at the promoter region of HOXC8 was detected for: BPTF at loci ˗1.4, +2, and +3 kb (P < 0.05); RBBP5 at loci ˗1.4, +2, +3, and 0 kb (P < 0.05); WDR5 at loci ˗1.4 and +2 kb (P < 0.05); and MLL at loci ˗1.4 and +2 kb (P > 0.05).
After siRNA-ASH2L treatment of THP-1 cells, decreased activity at the promoter region of HOXC8 was detected for: BPTF at loci ˗1.4 and +3 kb (P < 0.05); and RBBP5 at loci ˗1.4 and +3 kb (P < 0.05); however, the activities of WDR5 and MLL did not change at the four loci of the HOXC8 promoter (P > 0.05) (Supplemental Figure 5).
Discussion
HOX genes are widespread among organisms and can be divided into four clusters phylogenetically: A, B, C, and D. Previous studies have shown that HOX expression is closely related to the development of tumors. In normal tissues, the expression of HOX (which is only usually expressed in embryonic tissues) may lead to the development of tumors.7 The HOXC8 cluster is located on chromosome 12, and its subgroup HOXC8 is a type of evolutionarily highly conserved developmental regulatory gene that regulates the proliferation, apoptosis, migration, and differentiation of cells by regulating many genes at the transcriptional level. There is growing evidence that HOXC8 plays an important role in the development and progression of human malignancies, but the related mechanism is unclear.8–10
The MLL family of proteins is a class of methyltransferases specific for H3K4 whose methyltransferase activity depends on a conserved SET domain at the C-terminus. In MLL-rearranged acute leukemia, MLL fusion protein cannot directly activate downstream target genes by modifying the promoter region. Therefore, it is highly likely that there are other regulatory proteins in the promoter region of the target gene, or the regulatory protein plays a synergistic regulatory role by binding to the MLL gene. MLL family protein is different from other methyltransferases with a SET domain because its function requires a number of intrinsic auxiliary proteins, including WDR5, RBBP5, ASH2L and DPY30 to form a functional complex (COMPASS) to complete the methylation modification process.4,10 How this complex performs methylation modification remains in dispute. In recent years, research has focused on the role of ASH2L because ASH2L is an important and complex regulatory protein, and it has binding sites that can easily bind to other proteins. Moreover, ASH2L can bind to the promoter region of HOXC8, initiate the movement of nucleosomes and thereby activate the target gene.11–15 Based on these characteristics, we considered that ASH2L is likely to be involved in the transcriptional activation of the HOXC8 gene in MLL-rearranged acute leukemia.
To explore whether there are interactions between ASH2L and HOXC8 in MLL-rearranged acute leukemia, this study for the first time used siRNA transfection techniques to specifically interfere with the expression of ASH2L in RS4:11 and THP-1 MLL-rearranged acute leukemia cell lines. The results showed that the transfection efficiency was greater than 90%. We further studied the gene expression levels of ASH2L and HOXC8 after transfection, and the results showed significantly downregulated expression of both ASH2L and HOXC8.
To investigate whether ASH2L causes a decrease of H3K4-me3 in the HOXC8 promoter region, we next used ChIP-qRT-PCR to study the acute leukemia cells with two MLL genes rearranged after siRNA-ASH2L treatment. The H3K4-me3 levels at loci 0 kb (transcription start site in the promoter region of HOXC8), ˗2 and ˗3 kb (both upstream of the start site), and 1.4 kb (downstream of the start site) were analyzed. The results showed that, in RS4:11 cells after ASH2L interference, modification of H3K4-me3 at locus ˗3 kb upstream of the start site of the promoter region of HOXC8 was remarkably reduced. In THP-1 cells, modification of H3K4-me3 at loci ˗3 and ˗2 kb in the promoter region and locus 1.4 kb in the first intron of HOXC8 was significantly reduced (P < 0.05). These results confirmed our original hypothesis that ASH2L plays an important role in the regulation of histone methylation of acute leukemia with MLL rearrangement. ASH2L may directly or indirectly affect the expression of the target gene HOXC8 via interaction with MLL protein, increasing the H3K4-me3 level in the promoter region of HOXC8 and thereby activating HOXC8 expression. Chromatin remodeling is a recently described concept and the mechanism involved in chromatin remodeling is complex. The nucleosome slide along the chromatin, changing its structure from a tight to a loosened structure. The loosened structure may facilitate the binding of various transcription factors to the regulatory motif, thereby activating the gene.16–18 Chromatin remodeling is not an independent mechanism, but is regulated by multiple proteins, and is closely related to histone modifications. we thus further tested the binding between MLL, WDR5, and RBBP5 in the promoter region of HOXC8 and maximum nucleosome remodeling factor subunit PHD finger transcription factor (bromodomain and PHD finger transcription factor, BPTF) after siRNA-ASH2L interference. The results showed that after siRNA-ASH2L treatment of RS4:11 cells, decreased levels of BPTF and RBBP5 were detected at loci ˗1.4, +2, +3, and 0 kb in the promoter region of HOXC8 (P < 0.05); and MLL and WDR5 showed decreased activity at loci ˗1.4 and +2 kb in the promoter region of HOXC8 (P < 0.05). However, after siRNA-ASH2L treatment of THP-1 cells, BPTF and RBBP5 showed decreased activity at loci ˗1 and +3 kb in the promoter region of HOXC8 (P < 0.05); while no significant changes in WDR5 and MLL levels were observed for any of the four loci of HOXC8 (P > 0.05). This is possibly because the cell viability of THP-1 cells was relatively low compared with RS4:11 cells, thus leading to less reliable results. However, taken together, our results show that decreased expression of H3K4-me3 in the promoter region of HOXC8 was not directly due to the silencing of ASH2L.
In summary, our results provide insight into the association between histone methylation of the HOXC8 gene and MLL-rearranged acute leukemia. ASH2L may activate HOXC8 expression via interaction with MLL, WDR5, and RBBP5 of the MLL methylase complex, as well as BPTF protein, which modifies histone methylation and remodeling of the chromatin structure. Identification of ASH2L as a component of the MLL complex required for specific regulation of H3K4-me3 provides insight into the molecular mechanism of histone modification. Future genetic and biochemical studies on ASH2L and histone modification will be instrumental in defining the biological role of H3K4 methylation during development and differentiation.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (81370656), “Liu Da Ren Cai Gao Feng” of Jiangsu Province (number 2014-WSN-009).
Author Contributions
Yu-Jie Wu and Li-xia Li designed the research Lu Liu, Si-shu Zhao, and Qiu Hai-rong performed the research; Hui Wang analyzed the data; Yu-Jie Wu and Li-xia Li wrote the paper. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work. Yu-Jie Wu and Li-xia are co-first authors.
Disclosure
The authors report no conflicts of interest relating to this work.
References
1. Kohlmann A, Schoch C, Dugas M, et al. New insights into MLL gene rearranged acute leukemias using gene expression profiling: shared pathways, lineage commitment, and partner genes. Leukemia. 2005;19(6):953–964. doi:10.1038/sj.leu.2403746
2. Balgobind BV, Zwaan CM, Pieters R, et al. The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia. 2011;25(8):1239–1248. doi:10.1038/leu.2011.90
3. Swerdlow SH, Campo E, Hams NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon France: IARC press; 2008.
4. Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi:10.1146/annurev-biochem-051710-134100
5. Butler JS, Qiu YH, Zhang N, et al. Low expression of ASH2L protein correlates with a favorable outcome in acute myeloid leukemia. Leuk Lymphoma. 2017;58(5):1207–1218. doi:10.1080/10428194.2016.1235272
6. Pan JL, Xue YQ, Li JY, et al. Translocation t(11;19)(p23;p13) in a patient with therapy-related acute myeloid leukemia following Razoxane treatment for Psoriasis. J Leukemia Lymphoma. 2013;12(7):288–290.
7. Park S, Osmers U, Raman G, et al. The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression. Biochemistry. 2010;49(31):6576–6586. doi:10.1021/bi1009387
8. Li Y, Zhang M, Chen H, et al. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res. 2010;70(20):7894–7904. doi:10.1158/0008-5472.CAN-10-1675
9. Li Y, Guo Z, Chen H, et al. HOXC8- dependent cadherin 11 expression facilitates breast cancer cell migration through trio and rac. Genes Cancer. 2011;2(9):880–888. doi:10.1177/1947601911433129
10. Waltregny D, Alami Y, Clausse N, et al. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate. 2002;50(3):162–169. doi:10.1002/(ISSN)1097-0045
11. Chen Y, Wan B, Wang KC, et al. Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding. EMBO Rep. 2011;12(8):797–803. doi:10.1038/embor.2011.101
12. Tan CC, Walsh MJ, Gelb BD. Fgfr3 is a transcriptional target of Ap2delta and Ash2l-containing histone methyltransferase complexes. PLoS One. 2009;4(12):e8535. doi:10.1371/journal.pone.0008535
13. Xiao Y, Bedet C, Robert VJ, et al. Caenorhabditis elegans chromatin -associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc Natl Acad Sci USA. 2011;108(20):8305–8310. doi:10.1073/pnas.1019290108
14. Kawabe Y, Wang YX, McKinnell IW, et al. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11(3):333–345. doi:10.1016/j.stem.2012.07.001
15. Shinsky SA, Monteith KE, Viggiano S, et al. Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J Biol Chem. 2015;290(10):6361–6375. doi:10.1074/jbc.M114.627646
16. Koludrovic D, Laurette P, Strub T, et al. Chromatin-remodelling complex NURF is essential for differentiation of adult melanocyte stem cells. PLoS Genet. 2015;11(10):e1005555. doi:10.1371/journal.pgen.1005555
17. Xu B, Cai L, Butler JM, et al. The chromatin remodeler BPTF activates a stemness gene-expression program essential for the maintenance of adult hematopoietic stem cells. Stem Cell Rep. 2018;10(3):675–683. doi:10.1016/j.stemcr.2018.01.020
18. Li H, Ilin S, Wang W, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442(7098):91–95. doi:10.1038/nature04802
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.