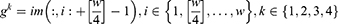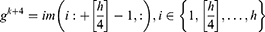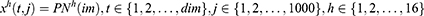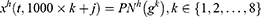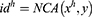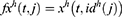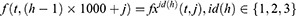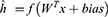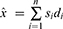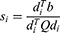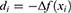Back to Journals » International Journal of General Medicine » Volume 15
Artificial Intelligence-Based Breast Cancer Diagnosis Using Ultrasound Images and Grid-Based Deep Feature Generator
Authors Liu H, Cui G, Luo Y, Guo Y, Zhao L, Wang Y, Subasi A , Dogan S, Tuncer T
Received 1 December 2021
Accepted for publication 11 January 2022
Published 1 March 2022 Volume 2022:15 Pages 2271—2282
DOI https://doi.org/10.2147/IJGM.S347491
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Haixia Liu,1 Guozhong Cui,2 Yi Luo,3 Yajie Guo,1 Lianli Zhao,4 Yueheng Wang,5 Abdulhamit Subasi,6,7 Sengul Dogan,8 Turker Tuncer8
1Department of Ultrasound, Cangzhou Central Hospital, Cangzhou, Hebei Province, 061000, People’s Republic of China; 2Department of Surgical Oncology, Cangzhou Central Hospital, Cangzhou, Hebei Province, 061000, People’s Republic of China; 3Medical Statistics Room, Cangzhou Central Hospital, Cangzhou, Hebei Province, 061000, People’s Republic of China; 4Department of Internal Medicine teaching and research group, Cangzhou Central Hospital, Cangzhou, Hebei Province, 061000, China; 5Department of Ultrasound, The Second Hospital of Hebei MedicalUniversity, Shijiazhuang, Hebei Province, 050000, People’s Republic of China; 6Institute of Biomedicine, Faculty of Medicine, University of Turku, Turku, 20520, Finland; 7Department of Computer Science, College of Engineering, Effat University, Jeddah, 21478, Saudi Arabia; 8Department of Digital Forensics Engineering, College of Technology, Firat University, Elazig, 23119, Turkey
Correspondence: Lianli Zhao, Department of Internal Medicine Teaching and Research Group, Cangzhou Central Hospital, Cangzhou, Hebei Province, 061000, People’s Republic of China, Tel +86 183 31125851, Email [email protected]
Purpose: Breast cancer is a prominent cancer type with high mortality. Early detection of breast cancer could serve to improve clinical outcomes. Ultrasonography is a digital imaging technique used to differentiate benign and malignant tumors. Several artificial intelligence techniques have been suggested in the literature for breast cancer detection using breast ultrasonography (BUS). Nowadays, particularly deep learning methods have been applied to biomedical images to achieve high classification performances.
Patients and Methods: This work presents a new deep feature generation technique for breast cancer detection using BUS images. The widely known 16 pre-trained CNN models have been used in this framework as feature generators. In the feature generation phase, the used input image is divided into rows and columns, and these deep feature generators (pre-trained models) have applied to each row and column. Therefore, this method is called a grid-based deep feature generator. The proposed grid-based deep feature generator can calculate the error value of each deep feature generator, and then it selects the best three feature vectors as a final feature vector. In the feature selection phase, iterative neighborhood component analysis (INCA) chooses 980 features as an optimal number of features. Finally, these features are classified by using a deep neural network (DNN).
Results: The developed grid-based deep feature generation-based image classification model reached 97.18% classification accuracy on the ultrasonic images for three classes, namely malignant, benign, and normal.
Conclusion: The findings obviously denoted that the proposed grid deep feature generator and INCA-based feature selection model successfully classified breast ultrasonic images.
Keywords: deep classification framework, deep neural network, grid-based deep feature generator, iterative feature selection, breast ultrasonography (BUS)
Introduction
Breast cancer is one of the leading causes of mortality in women worldwide,12 and 2.26 million women were diagnosed with breast cancer in 2020.3 According to the World Health Organization (WHO), this rate determined that breast cancer is the most common type of cancer among the women in the world. The fifth most common death rate (685.000) in the world is breast cancer. In the last 5 years, 7.8 million women were cured of breast cancer and survived.3,4 Breast cancer can occur in women of any age. However, it is more common in older ages. Early diagnosis is very important in breast cancer, as with all cancer types. Early diagnosis of breast cancer contributes to a reduction in the frequency of early deaths.5–7 Ultrasound imaging is a useful diagnostic technique for detecting and classifying breast abnormalities.8 Artificial intelligence (AI) is gaining popularity due to its superior performance in image-recognition tasks, and it is increasingly being used in breast ultrasonography (BUS). AI can provide a quantitative assessment by automatically identifying imaging data and making more accurate and reproductive imaging diagnoses.8–10 As a result, the use of AI in breast cancer detection and diagnosis is crucial.11 It may save radiologists time and compensate for certain beginners’ lack of experience and expertise.
We provided a machine-learning technique and proposed a strategy for analyzing benign and malignant breast tumor categorization in BUS images without requiring a priori tumor region-selection processing, reducing clinical diagnosis efforts while retaining good classification performance. This work aims to develop an AI-based diagnosis model of breast cancer detection using two-dimensional grayscale ultrasound images. AI techniques can perform well in identifying benign and malignant breast tumors. It has a potential to enhance diagnosis accuracy and minimize needleless biopsies of breast lesions in practice.
Several studies on breast cancer detection in the literature are presented. Qi et al12 proposed a breast cancer detection method using breast ultrasonography images. This method was based on deep neural networks. In their study, 8145 breast ultrasonography images were used. The accuracy rate for the two classes (malignant and non-malignant) is 90.13%. Eroglu et al13 presented a classification method based on CNN. The main purpose of the study is to detect breast cancer from BUS images, which consist of three classes (benign, malignant, and normal). In their study, 780 BUS images were used and 95.60% accuracy was achieved with the support vector machine classifier. Drukker et al14 proposed a computer-aided diagnosis system for breast cancer detection. They used 2409 sonographic images of 542 patients. Their method was based on Bayesian Neural Network and auto-assessment confidence level values. They obtained an AUC of 90.00% for 2 classes (benign and malignant). Nugroho et al15 conducted an approach based on a multilayer perceptron model for the classification of BUS images. Ninety-eight BUS images were used for the classification of breast cancer lesions. Nugroho et al15 obtained an accuracy of 87.79%. Babaghorbani et al16 applied a gray level co-occurrence matrix method for breast cancer classification. Twenty-five benign and 15 malignant images were utilized. They extracted 352 features from each image and reported an accuracy value of 85.00% for 2 classes (benign and malignant). Tan et al17 developed a computer-aided detection system for breast cancer detection. The main purpose of their study is to detect breast cancer using 3-D BUS images. They used 348 images of 238 patients and obtained patient-based sensitivity of 78.00%. Liao et al18 used a convolutional neural network for breast tumor detection using BUS images. In total, 256 images from 141 patients were collected with 130 benign and 126 malignant tumors. They obtained an accuracy rate of 92.95%. Liu et al19 presented a classification method for breast cancer detection using BUS images. They used iterated Laplacian regularization, which was used for dimensionality reduction. In total, 200 pathology images (100 malignant tumors and 100 benign masses) were selected to evaluate the proposed method. Takemura et al20 used log-compressed K-distribution for breast tumor detection. A total of 300 ultrasonic images (200 carcinomas, 50 fibroadenomas, and 50 cysts) were used to validate the proposed method. They reported an accuracy value of 100% using AdaBoost.M2 ensemble classifier. Joo et al21 developed an artificial neural network model for identifying breast nodules with BUS images. They used 99 malignant and 167 benign cases and obtained an accuracy of 91.40%. The works done before mostly achieved low accuracies with BUS images.
The novelties and contributions of our proposed framework are as follows:
Novelties
- A new deep feature generator (grid-based deep feature generator) is presented using pre-trained CNNs.
- A new grid-based deep feature generation framework is proposed using the widely used 16 CNNs. By using this framework, the optimal pre-trained CNNs are selected to solve computer vision problems.
Contributions
- Deep learning-based computer vision methods have yielded higher detection performance. However, the time complexities of deep models are very high because of their complex training phase. Transfer learning has been used to overcome this problem. Sixteen CNNs are used by applying transfer learning to create a lightweight and high-performing method. Moreover, CNNs have variable classification ability. In order to select the best-performed CNNs for the interesting computer vision problem, a new deep learning framework is presented. Moreover, local features are very meaningful to yield high performance. Therefore, patch-based deep models have attained high classification performance such as vision transformers and multilayer perceptron mixers. However, patch/exemplar-based models generate huge-sized features. A new grid-based deep feature generator is proposed using 16 pre-trained CNN models to utilize the effectiveness of the patch/exemplar-based models with fewer features. In this way, problems are solved by applying the proposed framework by the optimal deep feature generator selection and attaining high accuracy using fewer features (less complex feature generation procedure).
The presented grid-based deep feature generator model is used to create a cognitive machine learning model. Each phase of this model is designed cognitively, and it can select the optimal pre-trained CNN models, which use an iterative feature selector and a deep classifier to attain maximum performance. Finally, a BUS dataset is selected to test the performance of our proposed framework, and we have achieved 97.18% accuracy on this dataset.
Materials and Methods
Material
We utilized the publicly available BUS image dataset collected from Behaye Hospital and the dataset is freely available at the web site.22 The ethics committee of Behaye Hospital approved the study protocol. The data collected at baseline include breast ultrasound images from 600 female patients in ages between 25 and 75 years. This data were collected in 2018. The LOGIQ E9 ultrasound system and the LOGIQ E9 Agile ultrasound system were utilized in the scanning procedure. These devices are often utilized in high-end imaging for radiology, cardiology, and vascular applications. They generate images at a resolution of 1280*1024. The transducers on the ML6-15-D Matrix linear probe are 1–5 MHz. The dataset consists of 780 images with an average image size of 500×500 pixels. A BUS image dataset has been used to denote the comparative success of the proposed grid-based deep feature generator framework. This dataset is heterogeneous, and there are 133 normal, 437 benign, and 210 malignant images. The whole images are grayscale. The data were in DICOM format but converted into PNG format using a DICOM converter program. The BUS dataset contains three categories: normal, benign, and malignant. The total number of images acquired at the start was 1100. The duplicated images were eliminated. Furthermore, Baheya Hospital radiologists evaluated and corrected the inaccurate annotation. To remove unnecessary and insignificant borders from the images, all of them were cropped to different sizes. The image annotation is placed in the image caption. After preprocessing, the number of BUS images in the dataset was decreased to 780. The original images include irrelevant information that will not be used for mass categorization. Furthermore, they may have an impact on the training process’s outcomes. To make the ultrasound dataset useful, ground truth (image boundary) is done. For each image, a freehand segmentation is generated.22
Proposed Method
A new grid-based deep feature generator is proposed in this work, and a new computer vision framework is employed by applying the proposed generator. The main purpose of our feature generator is to attain high classification ability like exemplar-based deep models, but exemplar/patch-based deep models have a complex time burden. The used ultrasonic image is divided into lines and columns to decrease the complexity of the exemplar feature generation without decreasing performance. For instance, by using a 5×5 sized grid in an exemplar model, 25 exemplars are obtained. The feature generator should extract features from the generated 25 exemplars. However, by using our proposed model, 10 grids are obtained a 5×5 sized mask. The other problem of the deep learning-based models is to choose the most appropriate network to solve the problem. Therefore, many researchers have used the trial and error method to find the best model to solve their problems. In this study, we proposed a framework by using 16 CNNs. The presented framework generates an error vector used to choose the best model(s). In this work, the proposed grid-based feature generator is imported into this framework to achieve high accuracy for this problem. The proposed framework uses INCA23 to choose the most appropriate feature vector. The deep neural network is deployed to obtain results. The graphical outline of the proposed method is shown in Figure 1.
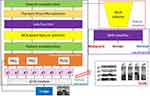 |
Figure 1 Graphical outline of the proposed grid-based deep framework. |
In the proposed framework, ultrasonic images are divided into grids, and eight grids (g1, g2, …, g8) are obtained. In the deep feature generation phase, the fully connected layer of each pre-trained model is used to feature extraction, and 9000 features are extracted from each ultrasonic image after the feature merging step. The top 1000 features of the generated 9000 features are chosen using NCA24 feature selector. The misclassification rate of each pre-trained model is calculated using an SVM25,26 classifier with 10-fold cross-validation. The SVM classifier is utilized as a loss function in this framework. By using the calculated loss values, the best pre-trained models are selected according to the computer vision problem. The final hybrid deep model for the ultrasonic image classification problem is shown in Figure 2. Figure 2 denotes the proposed grid-based deep transfer learning framework to find an appropriate model for medical image analysis problem. Furthermore, the proposed framework has three main phases (feature extraction, feature selection, and classification) that are denoted in Figure 1. Moreover, the pseudocode of this model is given in Algorithm 1.
Algorithm 1 Pseudocode of the Optimal Model Input: Ultrasound image dataset. Output: Results. 00: Load image dataset. 01: Read each image ultrasound dataset. 02: Divide grids to each image. Details of this step is given in Feature Extraction section. 03: Extract deep features from each grid and image using the pre-trained network. 04: Generate three feature vectors. 05: Choose the most informative 1000 features from each pre-trained network. 06: Merge these features and obtain final feature vector with a length of 3000. 07: Apply INCA selector to these 3000 features. 08: Forward the selected features to DNN classifier.
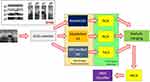 |
Figure 2 The created best model for the used ultrasonic image dataset using our proposed grid-based deep transfer learning framework. |
In testing phase, features are generated from images using the chosen optimal pretrained networks namely ResNet101, MobilNetv2 and EfficientNetb0. By deploying these three networks, three feature vectors with a length of 9000 have been created. Indexes of the most informative features are stored. By deploying these indexes, the most valuable features have been chosen. By using these indexes, there is no need to implement NCA and INCA in the testing phase. These features are classified by deploying a DNN classifier. More details about these phases are given in this section.
Feature Extraction
The most complex phase of the proposed grid-based deep transfer learning framework is the feature extraction since the feature generation directly affects the classification ability of the learning model. The proposed grid-based deep feature generator model selects features two times by using NCA and loss values to extract the most appropriate features to solve the classification problem. Steps of the presented grid-based deep feature generation model are given below.
Step 1: Create eight grids from the image.
where 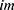 is the used ultrasonic image,
is the used ultrasonic image, 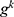 represents kth grid, and by using Equations (1) and (2), eight grids are generated,
represents kth grid, and by using Equations (1) and (2), eight grids are generated,  defines the width of the image and
defines the width of the image and 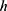 represents the height of the image. Hence, vertical and horizontal grids have been created. By employing Eqs. (1) and (2), eight grids have been generated.
represents the height of the image. Hence, vertical and horizontal grids have been created. By employing Eqs. (1) and (2), eight grids have been generated.
Step 2: Generate features from grids and original ultrasonic images using 16 pre-trained networks, which are ResNet18,27 ResNet50,27 ResNet101,27 DarkNet19,28 MobileNetV2,29 DarkNet53,28 Xception,30 EfficientNetbo,31 ShuffleNet,32 DenseNet201,33 Inceptionv3,34 InceptionResNetV2,35 GoogleNet,36 AlexNet, VGG1637 and VGG19.37 In this respect, this framework can be an extendable model, and more pre-trained networks can be added to generated features. These deep feature generators are selected by applying the proposed grid-based deep framework. The used pre-trained networks were trained on the ImageNet dataset. This dataset contains million of images with 1000 classes. Hence, each pre-trained network generates 1000 features. To generate these features, the last fully connected of these networks have been utilized.
where 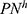 defines the used hth pre-trained network,
defines the used hth pre-trained network, 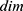 is the number of the used ultrasonic images,
is the number of the used ultrasonic images, 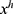 is hth generated feature vector with a length of 9000 and it is created from the original image and grids of the image. By applying Equations (3) and (4), 9000 features are generated using each pre-trained model. Equations (3) and (4) define feature extraction and merging phases together.
is hth generated feature vector with a length of 9000 and it is created from the original image and grids of the image. By applying Equations (3) and (4), 9000 features are generated using each pre-trained model. Equations (3) and (4) define feature extraction and merging phases together.
Step 3: Reduce the dimension of the extracted feature vectors ( ) deploying the NCA selector.
) deploying the NCA selector.
where, 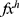 are selected features with a length of 1000. Equations (5) and (6) have been used to choose the most informative 1000 features from the generated 9000 features. The most informative/meaningful features are chosen using
are selected features with a length of 1000. Equations (5) and (6) have been used to choose the most informative 1000 features from the generated 9000 features. The most informative/meaningful features are chosen using 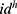 (qualified indexes according to the generated weights).
(qualified indexes according to the generated weights).
Step 4: Calculate misclassification rates of each feature vector ( ) deploying SVM classifier with 10-fold cross-validation. Herein, 16 misclassification rates are generated.
) deploying SVM classifier with 10-fold cross-validation. Herein, 16 misclassification rates are generated.
Step 5: Choose the best three pre-trained model using generated 16 loss values.
Step 6: Merge the 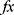 s to calculate the final feature vector.
s to calculate the final feature vector.
Herein, the best three feature vectors (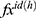 ) are chosen using loss values. In this work, the selected best pre-trained models for feature extraction are ResNet101, MobileNetV2, and EfficientNetb0. The created final vector (
) are chosen using loss values. In this work, the selected best pre-trained models for feature extraction are ResNet101, MobileNetV2, and EfficientNetb0. The created final vector (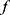 ) has 3000 features. In the feature selection phase, INCA is deployed to choose the best feature combination, and details of the feature selection are explained in Section B.
) has 3000 features. In the feature selection phase, INCA is deployed to choose the best feature combination, and details of the feature selection are explained in Section B.
Feature Selection
INCA is an iterative and developed version of the NCA, and it was proposed by Tuncer et al in 2020.23 INCA uses a loss function to select the best features, and it has an iterative structure. It is a parametric feature selection method. Users can define the initial value of the loop, the final value of the loop, and the loss function. Generally, a classifier has been utilized as a loss function. Loop range is defined to decrease the time complexity of the INCA. The initial value of the loop value is set to 100, and End value of the loop value is set to 1000, and third degree (Cubic) SVM with 10-fold cross-validation is utilized in the loss function. By using these parameters, the best features are selected from the generated 3000 features. The length of the selected best feature vector is found as 980.
Classification
The last phase is the classification of the proposed grid-based deep learning model.38 A deep neural network (DNN) is a form of artificial neural network (ANN) with two or more hidden layers. Because gradient computation of functions is required, the used DNN is a backward network that uses scaled conjugate gradient (SCG) for learning. The SCG algorithm employs the steepest descent direction. During the DNN implementation, the initial weights are randomly assigned, and 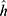 (input of hidden layers) are calculated using Eq. 8.
(input of hidden layers) are calculated using Eq. 8.
where W is assigned weights, x shows inputs, and f is the activation function. The weights are then recalculated using the backpropagation approach. In this phase, SCG is used, which is the steepest optimization approach, and it utilizes orthogonal vectors to minimize error. The mathematical notation of SCG is given in Eqs. 9–11.
where 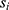 is multiplier, d is orthogonal vector and x is input. Weights are recalculated using this optimization method. To evaluate the efficacy of the proposed feature extraction and selection framework, the selected 980 features are fed into the SCG-based three hidden layered DNN. There is currently no standard method for building an ideal deep learning model with an adequate number of layers and neurons in each layer. As a result, we experimentally developed the DNN through several trials. In each experiment, we carefully tuned the number of hidden layers, the number of nodes generating the layer for each hidden layer, the number of learning steps, the learning rate, momentum, and the activation function. We used a SCG optimization approach for the backpropagation method and adjusted the learning rate to 0.7, momentum to 0.3, and batch size to 100. To find the remaining DNN hyperparameters, we computed the classification accuracy using 10-fold cross-validation for each manual formation.39 This method is repeated for various sizes of hidden layer representations. Following this laborious manual procedure, the best classification result is obtained with a DNN composed of three hidden layers of 400, 180, and 40 nodes, respectively. In this study, the scaled conjugate gradient is used as an optimizer. As an activation function, the tangent sigmoid is used. Furthermore, batch normalization is used in the model.
is multiplier, d is orthogonal vector and x is input. Weights are recalculated using this optimization method. To evaluate the efficacy of the proposed feature extraction and selection framework, the selected 980 features are fed into the SCG-based three hidden layered DNN. There is currently no standard method for building an ideal deep learning model with an adequate number of layers and neurons in each layer. As a result, we experimentally developed the DNN through several trials. In each experiment, we carefully tuned the number of hidden layers, the number of nodes generating the layer for each hidden layer, the number of learning steps, the learning rate, momentum, and the activation function. We used a SCG optimization approach for the backpropagation method and adjusted the learning rate to 0.7, momentum to 0.3, and batch size to 100. To find the remaining DNN hyperparameters, we computed the classification accuracy using 10-fold cross-validation for each manual formation.39 This method is repeated for various sizes of hidden layer representations. Following this laborious manual procedure, the best classification result is obtained with a DNN composed of three hidden layers of 400, 180, and 40 nodes, respectively. In this study, the scaled conjugate gradient is used as an optimizer. As an activation function, the tangent sigmoid is used. Furthermore, batch normalization is used in the model.
Results
A simple configured computer has been used to implement the proposed grid-based deep learning model. The used computer has 16 GB memory, an intel i7 7700 processor with 4.20 GHz clock, 1TB disk, and Windows 10.1 Professional operating system. The proposed grid-based deep learning model has been implemented by MATLAB 2020b programming tool. Firstly, we imported pre-trained networks to MATLAB using Add-Ons, and then our proposal was implemented using m-files. Pre-trained networks have been used with default settings. Any fine-tuning model has not been used on the pre-trained networks. Furthermore, any parallel programming method has not been used since transfer learning has been used to generate deep features.
We used accuracy, recall, precision, F1-score, and geometric mean to evaluate the performance of the proposed grid-based deep learning approach. The obtained confusion matrix is presented in Table 1. In this confusion matrix (see Table 1), true predicted and false predicted values are denoted. Moreover, class by a class recall, precision, and F1-scores are demonstrated in Table 2. As shown in Table 2, the proposed method reached over 96% for all performance metrics and yielded 97.18% classification accuracy. DNN classifier was used 10-fold cross-validation to achieve these results. Therefore, fold-wise accuracies are denoted in Figure 3. Figure 3 demonstrated that our proposal yielded 100% classification accuracy on both the third and seventh folds. The worst classification accuracy was calculated as 85.90% for the first fold.
 |
Table 1 The Generated Confusion Matrix |
 |
Table 2 Overall Results of the Proposed Grid-Based Deep Learning Model on the Used Ultrasonic Image Dataset |
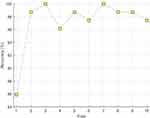 |
Figure 3 Fold-wise accuracies of the grid-based deep learning model on the used dataset. |
Discussion
In this work, we proposed a novel grid-based deep learning framework to attain high classification accuracy for breast cancer detection. Our proposed framework is a parametric framework, where 16 transfer learning methods are used to generate deep features. Moreover, eight grids are utilized. The top three pre-trained models are used to create a feature vector in the feature extraction phase. INCA was used to choose the best feature vector, and it selected 980 features as the best feature vector. In the classification phase, DNN is deployed with 10-fold cross-validation. According to the results, our proposed framework achieved 97.18% accuracy without using image augmentation method. The proposed grid-based model uses three feature selection methods. The first two feature selection methods are used in feature extraction: INCA and loss value generation based on top feature vectors’ selection. In order to select top feature vectors, Cubic SVM was deployed, and the calculated accuracy rates (1-loss) are tabulated in Table 3.
 |
Table 3 Accuracies of the Used 16 Pre-Trained Networks Using Cubic SVM with 10-Fold Cross Validation |
Table 3 demonstrates individual grid results of the used pre-trained networks using Cubic SVM, and we highlighted the selected networks using bold font type. Our main aim is to increase the performance (the best accuracy is 90.38%, according to Table 3) of the breast cancer detection using BUS images. Therefore, we merged these feature vectors and applied INCA to these features. Misclassification rates and the number of features selected by INCA are shown in Figure 4. According to Figure 4, 980 features are selected to attain maximum classification accuracy. By using 980 features, the proposed model reached 93.59% accuracy deploying Cubic SVM. Furthermore, the feature concatenation and INCA process increase the maximum accuracy rate from 90.38% to 93.59%.
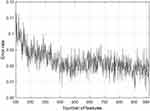 |
Figure 4 Number of features and misclassification rate (error rate) of the INCA for this work. |
The last phase of the grid-based deep learning model is classification. Cubic SVM was used to calculate error values to select the most informative features. In order to increase classification performance, a deep neural network (DNN) is used to increase the classification ability of proposed approach. According to Table 2, DNN attained 97.18% classification accuracy. This classifier (DNN) increased the classification ability of the proposed model approximately 3.6%.
These results clearly indicate that our grid-based model is a cognitive deep image classification model. The comparison with the state-of-the-arts is tabulated in Table 4 for the same dataset.
 |
Table 4 Comparison with the State-of-the-Arts Using the Same BUS Dataset |
Table 4 denotes that most of the works used deep learning models to achieve high classification rates, and some methods are used augmentation to attain high classification performance. Our model reached the best scores among these works (see Table 4).
In computer vision applications, CNNs have generally been used to attain high classification results and there are many CNNs in the literature. These models have individual success rates on the image databases. This research proposed a general deep framework to solve the breast cancer classification problem. Therefore, feature extraction capabilities of 16 pre-trained networks have been tested on the used dataset. Fixed-size patch-based models have been used to generate local deep features (comprehensive features). However, this approximation is a complex approximation. To generate local deep features and decrease the time, grid division has been presented. This model is an explainable image classification model since it chooses the best/most suitable models for solving image classification problems. In this respect, this model is a self-organized deep feature extraction model. DNN (deep neural network) has been applied to features and achieved higher classification performance than other state-of-the-art methods. The important points of this work are given as follows.
- Exemplar feature generators/deep models have attained high classification performances, but their time complexity is high. A grid-based model is proposed to decrease the time complexity of the exemplar model without decreasing the classification performance.
- A novel deep image classification framework is proposed to choose the most appropriate pre-trained networks (CNNs).
- We created an ultrasonic image classification method by using ResNet101, EfficientNetb0, and MobileNetV2 according to the results of our framework.
- Any augmentation model has not been used to increase classification accuracy.
- The proposed grid-based deep learning model is a cognitive ultrasonic image classification method for breast cancer detection.
- Our grid-based model outperforms (see Table 4).
- Our proposed framework can be used to solve other computer vision/image classification problems in future studies.
- More and bigger datasets can be used to test the proposed approach.
Conclusion
Ultrasonic image classification is one of the hot research topics for biomedical engineering and computer sciences since many diseases can be diagnosed using ultrasonic medical images. Furthermore, intelligent medical applications can be used in near future to save the time of both breast cancer patients and medical professionals. Therefore, automated models have been widely presented in the literature, and the flagship of the automatic classification methods is deep learning since deep networks have achieved higher performances. Therefore, various deep learning networks have been proposed. The main problem of deep learning is to select the appropriate models to solve the given problem. Therefore, we proposed a new grid-based deep learning framework to select the best performing networks automatically to detect breast cancer utilizing ultrasonic image dataset. By using this dataset, ResNet101, MobileNetV2, and EfficientNetb0 are selected in the proposed framework to create the best classification method. The created model achieved 97.18% classification accuracy using 10-fold cross-validation.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Harris JR, Lippman ME, Veronesi U, Willett W. Breast cancer. N Engl J Med. 1992;327(5):319–328. doi:10.1056/NEJM199207303270505
2. Carlson RW, Allred DC, Anderson BO, et al. Breast cancer. J Natl Compreh Cancer Net. 2009;7(2):122–192. doi:10.6004/jnccn.2009.0012
3. WHO. Cancer. Available from: https://www.who.int/news-room/fact-sheets/detail/cancer.
4. WHO. Breast cancer. Available from: https://www.who.int/news-room/fact-sheets/detail/breast-cancer.
5. Goss PE, Ingle JN, Martino S, et al. A randomized trial of letrozole in postmenopausal women after five years of tamoxifen therapy for early-stage breast cancer. N Engl J Med. 2003;349(19):1793–1802. doi:10.1056/NEJMoa032312
6. AlShamlan NA, AlOmar RS, Almukhadhib OY, et al. Characteristics of breast masses of female patients referred for diagnostic breast ultrasound from a Saudi primary health care setting. Int J General Med. 2021;14:755. doi:10.2147/IJGM.S298389
7. Ranjkesh M, Hajibonabi F, Seifar F, Tarzamni MK, Moradi B, Khamnian Z. Diagnostic value of elastography, strain ratio, and elasticity to B-mode ratio and color Doppler ultrasonography in breast lesions. Int J General Med. 2020;13:215. doi:10.2147/IJGM.S247980
8. Cheng HD, Shan J, Ju W, Guo Y, Zhang L. Automated breast cancer detection and classification using ultrasound images: a survey. Pattern Recogn. 2010;43(1):299–317. doi:10.1016/j.patcog.2009.05.012
9. Fei X, Zhou S, Han X, et al. Doubly supervised parameter transfer classifier for diagnosis of breast cancer with imbalanced ultrasound imaging modalities. Pattern Recogn. 2021;120:108139. doi:10.1016/j.patcog.2021.108139
10. Nascimento CDL, Silva S, Silva T, Pereira W, Costa MGF, Costa CFF. Breast tumor classification in ultrasound images using support vector machines and neural networks. Res Biomed Eng. 2016;32:283–292. doi:10.1590/2446-4740.04915
11. Wu -G-G, Zhou L-Q, Xu J-W, et al. Artificial intelligence in breast ultrasound. World J Radiol. 2019;11(2):19. doi:10.4329/wjr.v11.i2.19
12. Qi X, Zhang L, Chen Y, et al. Automated diagnosis of breast ultrasonography images using deep neural networks. Med Image Anal. 2019;52:185–198. doi:10.1016/j.media.2018.12.006
13. Eroğlu Y, Yildirim M, Çinar A. Convolutional Neural Networks based classification of breast ultrasonography images by hybrid method with respect to benign, malignant, and normal using mRMR. Comp Biol Med. 2021;133:104407. doi:10.1016/j.compbiomed.2021.104407
14. Drukker K, Sennett CA, Giger ML. Automated method for improving system performance of computer-aided diagnosis in breast ultrasound. IEEE Trans Med Imaging. 2008;28(1):122–128. doi:10.1109/TMI.2008.928178
15. Nugroho HA, Sahar M, Ardiyanto I, Indrastuti R, Choridah L. Classification of breast ultrasound images based on posterior feature. IEEE. 2016;1–4. doi:10.1109/IBIOMED.2016.7869825
16. Babaghorbani P, Parvaneh S, Ghassemi A, Manshai K. Sonography images for breast cancer texture classification in diagnosis of malignant or benign tumors. IEEE. 2010;1–4. doi: 10.1109/ICBBE.2010.5516073
17. Tan T, Platel B, Mus R, Tabar L, Mann RM, Karssemeijer N. Computer-aided detection of cancer in automated 3-D breast ultrasound. IEEE Trans Med Imaging. 2013;32(9):1698–1706. doi:10.1109/TMI.2013.2263389
18. Liao W-X, He P, Hao J, et al. Automatic identification of breast ultrasound image based on supervised block-based region segmentation algorithm and features combination migration deep learning model. IEEE J Biomed Health Inf. 2019;24(4):984–993. doi:10.1109/JBHI.2019.2960821
19. Liu X, Shi J, Zhou S, Lu M. An iterated Laplacian based semi-supervised dimensionality reduction for classification of breast cancer on ultrasound images. IEEE. 2014;4679–4682. doi:10.1109/EMBC.2014.6944668
20. Takemura A, Shimizu A, Hamamoto K. Discrimination of breast tumors in ultrasonic images using an ensemble classifier based on the AdaBoost algorithm with feature selection. IEEE Trans Med Imaging. 2009;29(3):598–609. doi:10.1109/TMI.2009.2022630
21. Joo S, Yang YS, Moon WK, Kim HC. Computer-aided diagnosis of solid breast nodules: use of an artificial neural network based on multiple sonographic features. IEEE Trans Med Imaging. 2004;23(10):1292–1300. doi:10.1109/TMI.2004.834617
22. Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data in Brief. 2020;28:104863. doi:10.1016/j.dib.2019.104863
23. Tuncer T, Dogan S, Özyurt F, Belhaouari SB, Bensmail H. Novel multi center and threshold ternary pattern based method for disease detection method using voice. IEEE Access. 2020;8:84532–84540. doi:10.1109/ACCESS.2020.2992641
24. Goldberger J, Hinton GE, Roweis S, Salakhutdinov RR. Neighbourhood components analysis. Advan Neural Inf Proc Syst. 2004;17:513–520.
25. Vapnik V. (1998) The Support Vector Method of Function Estimation. In: Suykens J.A.K., Vandewalle J. (eds) Nonlinear Modeling. Springer, Boston, MA. https://doi-org.ezproxy.utu.fi/10.1007/978-1-4615-5703-6_3
26. Vapnik V. The Nature of Statistical Learning Theory. Springer science & business media; 2013.
27. He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition; 2016:770–778.
28. Redmon J, Farhadi A. YOLO9000: Better, Faster, Stronger; 2017:7263–7271.
29. Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C. Mobilenetv2: Inverted Residuals and Linear Bottlenecks. 2018;4510–4520.
30. Chollet F. Xception: Deep Learning with Depthwise separable Convolutions. 2017;1251–1258.
31. Tan, M., & Le, Q. (2019, May). Efficientnet: Rethinking model scaling for convolutional neural networks. In International conference on machine learning (pp. 6105-6114). PMLR. http://proceedings.mlr.press/v97/tan19a/tan19a.pdf
32. Zhang X, Zhou X, Lin M, Sun J. Shufflenet: An Extremely Efficient Convolutional Neural Network for Mobile Devices; 2018;6848–6856.
33. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely Connected Convolutional Networks; 2017;4700–4708.
34. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the Inception Architecture for Computer Vision. 2016;2818–2826.
35. Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, Inception-Resnet and the Impact of Residual Connections on leArning. 2017.
36. Szegedy C, Zaremba W, Sutskever I, et al. Intriguing properties of neural networks. arXiv preprint arXiv:13126199; 2013.
37. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556. 2014.
38. Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? arXiv preprint arXiv:14111792; 2014.
39. Ozyurt F, Tuncer T, Subasi A. An automated COVID-19 detection based on fused dynamic exemplar pyramid feature extraction and hybrid feature selection using deep learning. Comp Biol Med. 2021;132:104356. doi:10.1016/j.compbiomed.2021.104356
40. Moon WK, Lee Y-W, Ke -H-H, Lee SH, Huang C-S, Chang R-F. Computer‐aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks. Comp Methods Prog Biomed. 2020;190:105361. doi:10.1016/j.cmpb.2020.105361
41. Byra M, Jarosik P, Szubert A, et al. Breast mass segmentation in ultrasound with selective kernel U-Net convolutional neural network. Biomed Signal Proc Control. 2020;61:102027. doi:10.1016/j.bspc.2020.102027
42. Sadad T, Hussain A, Munir A, et al. Identification of breast malignancy by marker-controlled watershed transformation and hybrid feature set for healthcare. Appl Sci. 2020;10(6):1900. doi:10.3390/app10061900
43. Mishra AK, Roy P, Bandyopadhyay S, Das SK. Breast ultrasound tumour classification: a Machine Learning—Radiomics based approach. Expert Syst. 2021;38:e12713.
44. Muduli D, Dash R, Majhi B. Automated diagnosis of breast cancer using multi-modal datasets: a deep convolution neural network based approach. Biomed Signal Proc Control. 2021;71:102825.
45. Joshi RC, Singh D, Tiwari V, Dutta MK. An efficient deep neural network based abnormality detection and multi-class breast tumor classification. Multimedia Tools Appl. 2021;1–21. doi:10.1007/s11042-021-11240-0
46. Zhang G, Zhao K, Hong Y, Qiu X, Zhang K, Wei B. SHA-MTL: soft and hard attention multi-task learning for automated breast cancer ultrasound image segmentation and classification. Int J Comp Assisted Radiol Surg. 2021;1–7. doi:10.1007/s11548-021-02445-7
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.