Back to Journals » OncoTargets and Therapy » Volume 13
Up-Regulation of Hsa_circ_0008792 Inhibits Osteosarcoma Cell Invasion and Migration and Promotes Apoptosis by Regulating Hsa-miR-711/ZFP1
Authors Chen L, Shan Y, Zhang H , Wang H, Chen Y
Received 21 November 2019
Accepted for publication 2 March 2020
Published 11 March 2020 Volume 2020:13 Pages 2173—2181
DOI https://doi.org/10.2147/OTT.S239256
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Lu Chen, Yu Shan, Hengshuo Zhang, Hongliang Wang, Yunzhen Chen
Department of Spine Surgery, Qilu Hospital, Shandong University, Jinan, Shandong Province 250012, Republic of China
Correspondence: Hongliang Wang; Yunzhen Chen
Department of Spine Surgery, Qilu Hospital, Shandong University, Jinan, Shandong Province 250012, Republic of China
Email [email protected]; [email protected]
Objective: Circular RNA is a newly discovered non-coding RNA. It plays an important role in regulating gene expression, and may take part in tumor progression. This study aimed to investigate the functions of hsa_circ_0008792 in osteosarcoma regulation.
Methods: We identified a circular RNA, hsa_circ_0008792, by using bioinformatics to analyze the GSE96962 dataset. The capacities of migration and invasion were assessed by wound-healing assay and transwell Matrigel assay. The ratios of G0/G1, S, and G2/M phases in cell cycle and apoptosis were measured using flow cytometry.
Results: Hsa_circ_0008792 is expressed at low levels in osteosarcoma cells, and up-regulation of hsa_circ_0008792 could suppress osteosarcoma cell migration and invasion and promote apoptosis. This regulation is mediated by hsa-miR-711/ZFP1. The expression level of hsa_circ_0008792 showed no influence on cell cycle of osteosarcoma cells.
Conclusion: Osteosarcoma is suppressed by hsa_circ_0008792/hsa-miR-711/ZFP1 axis.
Keywords: circular RNA, microRNA, ZFP1, osteosarcoma, ceRNA network
Introduction
Osteosarcoma is the most common primary malignant bone tumor. It is the third largest malignant tumor causing death in adolescents worldwide.1,2 It is defined as the malignant mesenchymal cells that produce osteoid or immature bone.3,4
Although it has a lower morbidity rate than other cancers, osteosarcoma mainly occurs in adolescents and children and there is currently no effective therapy available, which places an increasing burden on society and the patients’ families. For osteosarcoma patients, the five-year survival rate is only approximately 60–70%.5 The survival rate of patients in advanced stage or with metastasis is only about 20%.6 Surgery combined with chemotherapy or radiotherapy is the main therapeutic strategy for osteosarcoma, but outcomes remain unsatisfactory.7 Thus, it is important and necessary to explore the etiology of osteosarcoma, determine its molecular mechanism, and develop new therapeutic strategies.
Circular RNA is a special endogenous non-coding RNA with a closed-loop structure. Circular RNA was once considered a by-product of functional translation; however, with the discovery of its functions, circular RNA has become increasingly important. The biological function of circular RNAs falls into five main categories—sponge effect, post-transcriptional regulation, rolling circle translation, circular RNA-derived pseudogenes, and splicing interference—and also varies by location, binding site, and acting mode.8 In recent years, increasing evidence has shown that circular RNAs play important roles in the development of cancers.9 Several circular RNAs have been shown to take part in regulation of proliferation, apoptosis, migration, and invasion in osteosarcoma, thereby affecting prognosis.10–13 It is important to identify more unknown circular RNAs that may be involved in carcinogenesis and cancer progression, especially in osteosarcoma.
MicroRNA is also a form of endogenous non-coding RNA (ncRNA); each one contains 20–24 nucleotides.14 They are always sponged to circular RNA. As classic non-coding RNAs, microRNAs are explored extensively, and many of them are associated with cancers.15–17
Zinc finger proteins (ZFPs) are the largest DNA-binding protein family; they are encoded by approximately 2% of human genes. They also have many other biological functions by which they regulate gene expression, including RNA packaging, transcriptional activation, protein folding, and assembly. Eight different classes of zinc finger motifs have been described. The research object of this study, zinc finger protein 1 (ZFP1), is a C2H2 zinc finger protein, and it can regulate sexual reproduction and virulence in Cryptococcus neoformans,18 but studies about neoplasm regulation have been rare.
In this study, we screened the Gene Expression Omnibus (GEO) database and chose a specific circular RNA, hsa_circ_0008792, which has lower expression in osteosarcoma than in healthy osteoblasts. Most circular RNAs are demonstrated as enhancers to osteosarcoma.12,19,20 Hence, it would be meaningful to identify new circular RNAs that act as suppressors to osteosarcoma, such as hsa_circ_0008792. In this study, the inhibitory effect of the hsa_circ_0008792/hsa-miR-711/ZFP1 axis on osteosarcoma was explored.
Materials and Methods
Cell Culture
Osteosarcoma cell lines MG63 and U2OS, osteoblast cell line hFOB1.19, and embryonic kidney cell line HEK293 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, US). All of the cell lines were cultured with the corresponding culture medium according to the instructions from ATCC in 5% CO2 at 37°C.
Quantitative Real-Time PCR
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA, US). cDNA was reversely transcribed using Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, US) in accordance with the manufacturer’s protocols. The expression level of RNAs was detected using quantitative real-time PCR (RT-qPCR) with Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific). The data were obtained and analyzed using the Mx4000 Multiplex Quantitative PCR System (Stratagene, La Jolla, CA, US) equipped with Version 3.0 software. All values were normalized to an endogenous glyceraldehyde-3-phosphate dehydrogenase (GADPH) control.
Western Blotting
Cells were lysed by RIPA buffer (Beyotime Biotechnology, Beijing, China) with protease inhibitor cocktail (20X), ultrasonication was performed and then cells were centrifuged at 13,000 rpm at 4°C for 30 min. The supernatant was collected and loading buffer (5X) was added. The sample was separated with SDS-PAGE by electrophoresis. Then, proteins were electrotransferred onto a PVDF membrane (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, US). The membrane was blocked with 5% BSA (Solarbio Life Sciences, Beijing, China), and incubated at 4°C overnight with ZFP1 primary antibody (Invitrogen, 1:1000). Membranes were washed with TBST three times (10 min per time), incubated with horseradish peroxidase-conjugated Rabbit secondary antibodies (1:10,000; Zhongshan Golden Bridge Biotechnology, Beijing, China), and then washed thrice with TBST. Protein bands were visualized with an electrochemiluminescence reagent (Advansta, Inc., Menlo Park, CA, US).
Wound-Healing Assay
Cells were cultured in six-well dishes (Corning Incorporated, Corning, NY, US) with RPMI-1640 with 10% fetal bovine serum (FBS) (GIBCO BRL, Grand Island, NY, US). When the cells grew to cover more than 90% of the area of the bottom of the dish, a scratch was performed with a 200-μL pipette tip to make a linear wound in the central area, and then the bottom of the dish was washed with phosphate-buffered saline (PBS). Migrated cells in wound areas were counted under a phase-contrast microscope after 48 h.
Matrigel Invasion Assay
Matrigel (BD Biosciences, San Jose, CA, US) was thawed at 4°C overnight and diluted with 1640 medium. A total of 60 μL diluted Matrigel was put into the upper chamber of a 24-well transwell (NEST Biotechnology, Wu Xi, Jiangsu, China) and gelled at 37°C for 30 min. Then, 2×104 cells were seeded on the Matrigel in the upper chamber with 1640, and the lower chamber was added into 1640 with 10% FBS. Non-invaded cells on the top of the transwell were scraped off with a cotton swab. The invasive cells that adhered to the lower surface of the filter were fixed with methanol and stained with crystal violet. Invaded cells were counted under a light microscope.
CCK8 Assay
A total of 2000 cells with 100 μL medium were seeded in 96-well dishes. The medium in each well was replaced with 100 μL serum-free medium containing 10 μL CCK8 (Beyotime Biotechnology, Beijing, China) and cultured for 2 h before detection. The absorbance OD450 was measured using a microplate reader.
Cell Cycle Assessment
A total of 0.1 million cells run through different treatments were seeded in a six-well plate. Propidium iodide (PI) (Solarbio Life Sciences, Beijing, China) was used for DNA staining. The samples were treated with 5 μL PI, and incubated for 15 min in the dark at room temperature. Then the fluorescence signals were detected using flow cytometry (BD BioSciences, San Jose, CA, US) and the data were processed using ModFit (Version 3.2, Verity Software House, Inc., Topsham, ME, US).
Annexin V/PI Assay
A total of 0.1 million cells run through different treatments were seeded in six-well dishes and cells were collected after 24 h. The process of PI and Annexin V-FITC (BD Pharmingen, San Diego, CA, US) staining was carried out according to the protocol supported by ThermoFisher Scientific. The ratio of apoptosis was detected using flow cytometry (BD BioSciences, San Jose, CA, US) and cells in right lower quadrant were regarded as apoptosis cells.
Dual Luciferase Reporter Assay
HEK-293 cells were transfected with wild-type or mutated circular RNA plasmids, and with miR-711 mimics or negative controls. After 24 h, a dual luciferase reporter assay was performed using a Dual Luciferase Reporter Assay System (Promega, Madison, WI, US). The firefly luciferase activity was normalized to Renilla luciferase activity to facilitate comparison.
Nude Mice Xenograft Model
Nude mice (four weeks old) were purchased from Model Animal Research Center of Nanjing University (Nanjing, Jiangsu, China). The animals were housed under standard conditions (24°C± 2°C, 20 relative humidity, 12 hr light/dark cycles) and with unlimited access to standard rodent maintenance feed (KEAO XIELI FEED Co., Ltd, Beijing, China) and water. Hygienic conditions were maintained by weekly cage changes. All animals were used in accordance with institutional guidelines and the current experiments were approved by the Use Committee for Animal Care of the institute. For tumor-bearing model experiments, the animals were divided into two groups with 3 animals each group, and 10 million newly digested cells (with blank vector and with stable over-expression of hsa_circ_0008792) were injected into the oxter region. After 28 days, the mice were sacrificed by anesthesia with pentobarbital, and the tumors were excised.
Statistics
For the results, the difference between two groups was evaluated by Student’s t-test. For comparisons among more than two groups, one-way analysis of variance (one-way ANOVA) was used for analysis. Statistical software SPSS 22.0 was used for statistical analysis.
Results
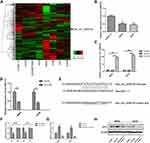
Hsa_circ_0008792 Was Low-Expressed in Osteosarcoma Cells
Through analysis of the GSE96964 microarray, we identified differentially expressed circular RNAs based on the criteria of fold change >2 and P value, and hsa_circ_0008792 was found to be expressed at low levels in osteosarcoma cell lines (Figure 1A). Subsequently, our results fully confirmed the down-regulation of hsa_circ_0008792 in osteosarcoma cell MG63 and U2OS relative to osteoblast cell hFOB1.19 (Figure 1B).
Hsa_circ_0008792 Could Regulate Hsa-miR-711/ZFP1 Expression
To investigate the biological effect of hsa_circ_0008792 in osteosarcoma cells, hsa_circ_0008792 over-expression plasmids were used for the transfection of MG63 and U2OS cells. The transfection efficacy was verified by q-PCR (Figure 1C), and up-regulation of hsa_circ_0008792 was found to decrease the expression of hsa-miR-711 (Figure 1D). The potential binding site between hsa_circ_0008792 and hsa-miR-711 was predicted by bioinformatics (TargetScan, http://www.targetscan.org/; circBank, http://www.circbank.cn/) (Figure 1E), and it was further verified by dual-luciferase reporter gene assay. The results showed lower luciferase intensity in MG63 and U2OS cells co-transfected with wild-type hsa_circ_0008792 and has-miR-711 mimics. However, there were no obvious changes in luciferase intensity in cells co-transfected with mutant-type hsa_circ_0008792 and hsa-miR-711 mimics (Figure 1F). The expression of ZFP1 could be regulated by either hsa_circ_0008792 over-expression or hsa-miR-711 mimics (Figure 1G and 1H). As shown in Figure 2, hsa_circ_0008792 could down-regulate the expression of hsa-miR-711 via sponge effect, and up-regulate the ZFP1 expression further.
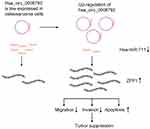 |
Figure 2 Diagrammatic sketch for mechanism of hsa_circ_0008792/hsa-miR-711/ZFP1 axis in osteosarcoma cell suppression. |
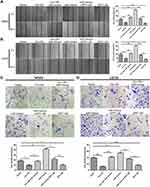
Up-Regulation of Hsa_circ_0008792 Inhibits Migration and Invasion in Osteosarcoma Cells Through Regulating Hsa-miR-711/ZFP1
Wound-healing assay revealed that over-expression of hsa_circ_0008792 markedly suppressed the capacity of migration in osteosarcoma cells. Hsa-miR-711 mimics would weaken the inhibitory effect from up-regulation of hsa_circ_0008792 and promote migration. But ZFP1 has an effect opposite that of hsa-miR-711. Over-expression of ZFP1 would decrease the capacity of migration and suppress the promotive effect of hsa-miR-711 (Figure 3A and B). As shown in Figure 3C and D, Matrigel invasion assay proved hsa_circ_0008792/hsa-miR-711/ZFP1 axis has a similar function in regulating osteosarcoma invasion. Combined with the result of Figure 1G, we drew the conclusion that up-regulation of hsa_circ_0008792 inhibited migration and invasion in osteosarcoma cells through regulating hsa-miR-711/ZFP1.
Up-Regulation of Hsa_circ_0008792 Suppresses Osteosarcoma Cells' Proliferation in vivo and in vitro
CCK8 assay revealed that over-expression of hsa_circ_0008792 markedly suppressed the proliferation rate of MG63 (Figure 4A) and U2OS (Figure 4B). The circ-OE group showed a lower proliferation rate than the control group. For the tumor-bearing model experiment, there are no mice found dead over the course of the study and the tumor weight in circ-OE group is smaller also (Figure 4C).
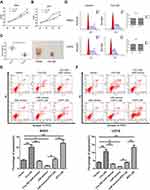
Hsa_circ_0008792 Suppresses Cell Proliferation via Promoting Cell Apoptosis Instead of Influencing Cell Cycle
Flow cytometry indicated that up-regulation of hsa_circ_0008792 could increase the rate of apoptosis in osteosarcoma cells through regulating hsa-miR-711/ZFP1, and the results were found to be statistically significant (Figure 4E and F), but no changes were visible in the cell cycle regardless of whether the expression of hsa_circ_0008792 was high or low (Figure 4D).
Discussion
The etiology and pathogenesis of osteosarcoma are currently not clear. As a very important regulator during gene expression and post-transcription modification, competing endogenous RNAs (ceRNA) network has attracted increasing attention.
Circular RNA is a key constituent part of ceRNA network and it has many functions, among which the “circRNA-miRNA-mRNA axis,” also called “miRNA sponge,” is the most frequently explored.19 This type of regulation is highly complex because of variation in circular RNAs and miRNAs and the variability of interactions between them.
An increasing number of studies have shown that circular RNA plays an important role in occurrence, development, and prognosis of malignant tumors. It is necessary and meaningful to clearly establish the functions of circular RNA in progression of osteosarcoma.
In this study, we focused on hsa_circ_0008792, which could interact with hsa-miR-711 to regulate the expression of ZFP1 and play an important role in the progression of osteosarcoma. The axis, hsa_circ_008792/hsa-miR-711/ZFP1 axis, is first found in this study, and it is also verified by experiments in this study. The binding site of hsa_circ_008792 and hsa-miR-711 was predicted by bioinformatics first and the sponge effect between them was also demonstrated by Dual luciferase reporter assay. Besides, hsa-miR-711 expression is down-regulated after up-regulation of hsa_circ_0008792. As for the downstream molecule, the expression of ZFP1 could be up-regulated via hsa_circ_0008792 over-expression or down-regulated via treating with hsa-miR-711 mimics in both mRNA and protein level.
It was first found that hsa_circ_0008792 is expressed at lower levels in osteosarcoma cells than in healthy osteoblast cells and that it acts as a cancer suppressor. As a kind of malignant neoplasm, osteosarcoma is featured in the rapid increase of number and distant metastasis, which are also the main reason for the shortened survival time of patients. Number increase is determined by proliferation and apoptosis while the capacity of metastasis is mainly influenced by migration and invasion. So it is very important and necessary to explore the effect of hsa_circ_0008792 on the above-mentioned activities in osteosarcoma cells, which may be contributed to clarify the growth and metastasis mechanism and corresponding regulation target better.
Up-regulation of hsa_circ_0008792 was found to decrease the osteosarcoma cells’ capacity for migration and invasion and increase apoptosis and these effects are regulated by the hsa_circ_0008792/hsa-miR-711/ZFP1 axis (Figure 2). Focusing on the molecular pathogenesis of osteosarcoma could provide a new therapeutic target for osteosarcoma. However, it will be better to investigate the biological behavior of osteosarcoma and the expression of circular RNAs, micro RNAs and relative proteins in vivo. It will contribute to a better understanding of the relationship between these ncRNAs and the significance of them in clinical work. It may be a limitation of this study that missing this part.
Some ncRNAs, like lncRNA and microRNA, can be detected in such body fluids as plasma, so ncRNAs could be used as biomarkers for diseases, which would allow a more prompt, less invasive diagnosis. It would be very meaningful to determine whether hsa_circ_0008792 is detectable in plasma or whether there is any obvious relationship between the expression of hsa_circ_0008792 and the clinical stage of osteosarcoma. This could facilitate accurate diagnosis, and should be the focus of future research.
Conclusion
Up-regulation of hsa_circ_0008792 inhibits invasion and migration and promotes apoptosis in osteosarcoma by regulating hsa-miR-711/ZFP1. The level of expression of hsa_circ_0008792 has no influence on the cell cycle.
Data Sharing Statement
The data in this study are available from the corresponding author upon reasonable request.
Ethics and Consent Statement
This study was approved by the ethics committee of Qilu Hospital and is numbered KYLL-2019(KS)-365. Non-retrospective ethical approval was obtained for the animal experiments conducted in the study. In addition, the authors confirmed that the tumor burden did not exceed the recommended dimensions and animals were anesthetized and sacrificed using acceptable methods.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; all took part in drafting or critically revising the article for important intellectual content; all gave final approval for the version to be published; and all agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
1. Simpson E, Brown HL. Understanding osteosarcomas. JAAPA. 2018;31(8):15–19. doi:10.1097/01.JAA.0000541477.24116.8d
2. Moore DDLH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92.
3. Fletcher CD, Unni KK, Mertens F. World Health Organization classification of tumours. Pathol Genet Tumours Soft Tissue Bone. 2002.
4. Arndt CA, Crist WM. Common musculoskeletal tumors of childhood and adolescence. N Engl J Med. 1999;341:342–352. doi:10.1056/NEJM199907293410507
5. Zhang H, Wang G, Ding C, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8(37):61687–61697. doi:10.18632/oncotarget.18671
6. Zhang Y, Meng W, Cui H. LncRNA CBR3-AS1 predicts unfavorable prognosis and promotes tumorigenesis in osteosarcoma. Biomed Pharmacother. 2018;102:169–174. doi:10.1016/j.biopha.2018.02.081
7. Bielack SS, Kempf-bielack B, Delling G, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20(3):776–790. doi:10.1200/JCO.20.3.776
8. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58. doi:10.1186/s12943-017-0630-y
9. Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi:10.1016/j.canlet.2018.03.035
10. Li JF, Song YZ. Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumour Biol. 2017;39(7):1010428317709991.
11. Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495(3):2369–2375. doi:10.1016/j.bbrc.2017.12.050
12. Li L, Guo L, Yin G, Yu G, Zhao Y, Pan Y. Upregulation of circular RNA circ_0001721 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-569 and miR-599. Biomed Pharmacother. 2019;109:226–232. doi:10.1016/j.biopha.2018.10.072
13. Jin H, Jin X, Zhang H, Wang W. Circular RNA hsa-circ-0016347 promotes proliferation, invasion and metastasis of osteosarcoma cells. Oncotarget. 2017;8(15):25571–25581. doi:10.18632/oncotarget.16104
14. Esquela-kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi:10.1038/nrc1840
15. Miao J, Wu S, Peng Z, Tania M, Zhang C. MicroRNAs in osteosarcoma: diagnostic and therapeutic aspects. Tumour Biol. 2013;34(4):2093–2098. doi:10.1007/s13277-013-0940-7
16. Nugent M. microRNA and bone cancer. Adv Exp Med Biol. 2015;889:201–230.
17. Zhang J, Yan YG, Wang C, Zhang SJ, Yu XH, Wang WJ. MicroRNAs in osteosarcoma. Clin Chim Acta. 2015;444:9–17. doi:10.1016/j.cca.2015.01.025
18. Fan CL, Han LT, Jiang ST, Chang AN, Zhou ZY, Liu TB. The Cys2His2 zinc finger protein Zfp1 regulates sexual reproduction and virulence in cryptococcus neoformans. Fungal Genet Biol. 2019;124:59–72. doi:10.1016/j.fgb.2019.01.002
19. Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. doi:10.1186/s12943-019-1007-1
20. Qi H, Sun Y, Jiang Y, Li X. Upregulation of circular RNA circ_0000502 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-1238. J Cell Biochem. 2018.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.