Back to Journals » Infection and Drug Resistance » Volume 11
Epidemiology of antibiotic resistance of blood culture in educational hospitals in Rasht, North of Iran
Authors Keihanian F, Saeidinia A, Abbasi K, Keihanian F
Received 24 March 2018
Accepted for publication 6 September 2018
Published 10 October 2018 Volume 2018:11 Pages 1723—1728
DOI https://doi.org/10.2147/IDR.S169176
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Fatemeh Keihanian,1 Amin Saeidinia,1,2 Keivan Abbasi,1 Faeze Keihanian2–5
1Faculty of Medicine, Guilan University of Medical Sciences, Rasht, Iran; 2Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran; 3Cardiology Department, Imam Reza Hospital, Mashhad, Iran; 4Cardiology Department, Ghaem International Hospital, Rasht, Iran; 5Pharmaceutical Research Division, Booali Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
Introduction: Septicemia is an emergency in medicine due to various dangers and lethality. In patients with septicemia, blood culture and other cultures should be done before antibiotic therapy. Determination of each isolated bacterium from blood culture and pattern of antibiotic sensitivity have an important role epidemiologically in a region and can assist physicians in the determination of primary antimicrobial agents. In this study, the researchers evaluated the schema of resistance of antibiotics in blood culture during the years 2014–2015 at eight educational hospitals in Rasht, North of Iran.
Materials and methods: This was a descriptive cross-sectional study. The researchers evaluated all blood cultures performed at four educational hospitals in Rasht. Demographic data were completed by a checklist, and antibiotic sensitivity antibiogram for all blood culture samples was done by diffusion agar (Kirby–Bauer method). Data were entered and analyzed in the SPSS software version 16, and the pattern of resistance, sensitivity, and intermediate antibiogram results was classified.
Results: From 3,283 cases, 225 subjects were positive (6.85%). Sixty-two percent of patients were male and others were female. The most common agent in the samples were Pseudomonas aeruginosa (66, 29.3%), coagulase-negative Staphylococcus (including Staphylococcus epidermidis and Staphylococcus saprophyticus; 50, 22.2%), coagulase-positive Staphylococcus aureus (29, 12.9%), and Escherichia coli (28, 12.4%). The most resistant antibiotic was ceftriaxone (44%), and the most sensitive antibiotic was ciprofloxacin (54%).
Conclusion: According to significant changes of microorganisms that cause sepsis and meaningful alterations of antibiogram findings, the current findings showed that choosing the best choice for empiric therapy should be corrected. The rate of positive samples in the current study was higher than the normal range. Consulting with a pharmacist for antibiotic prescription in hospitals and participation of physicians in antibiotic administration workshops could help decrease resistance.
Keywords: antibiotic resistance, blood culture, pattern of antibiograms, empiric therapy
Introduction
Infections of blood can be caused by multidrug resistance and can lead to more death and complications in hospitalized patients.1 For more than 50 years, the use of antibiotics has faced different alterations, such as change in kind and family of antibiotics, sensitivity, and resistance pattern to various antibiotics due to different causes such as emerging and re-emerging infectious disorders, over-the-counter use, and nonprescription utility of many antibiotics.2,3 Knowledge of trend in sensitivity and resistance patterns can help physicians and policy makers of every country and region to make better decisions on their approach to resolve probable resistance and general principles in their environment.4,5 Lack of national antibiotic resistance monitoring in developing countries will lead to unsuitable use in people and healthcare system workers.6 Development of infections and antibiotic resistance is a general concern around the world, and their incidence is globally increasing and can be different by region and according to geographic and epidemiological features.7,8 In hospitals and healthcare systems, infection by multidrug resistance can result in long-term hospitalization of patients, increase the rate of mortality, risk of other infections, and cost of the healthcare system.9 There are limited data on antibiotic resistance in general and central university hospitals of Iran. Therefore, in this investigation, the researchers analyzed the pattern of antibiotic resistance for different organisms at four educational high-referred hospitals of Rasht, North of Iran, during years 2014–2015, which can be a beneficial model for both clinicians in empirical therapy and policy makers.
Materials and methods
Study environment
This was a descriptive observational cross-sectional study. This study was performed on blood cultures requested at four educational referral hospitals of Rasht; Heshmat (cardiac referral center in Guilan Province), Poorsina (trauma referral center in Guilan province), Razi (general referral hospital in Guilan province), and Velayat (burning referral center in Guilan province) during February 2014–December 2015. Rasht is the center of Guilan province in the North of Iran with a population of 800,000. Guilan province is one of the Northern provinces of Iran, lying below the Caspian Sea, and has a population of nearly 2.5 million. There are some referral hospitals in Rasht, and the turnover of patients at these centers, which are educational and academic hospitals, is very high. Therefore, the importance of antibiotic resistance in this environment should be considered. These hospitals have covered different types of disorders and can be a good source of information of Guilan province’s status in antibiotic resistance.
Sample preparation
Blood culture of patients was kept in bottles at the laboratory for 7 weeks in a 37-degree incubator. Three blood samples for culture were obtained from every patient. Then, after 24 hours, subculture sampling was performed on MacConkey and blood agar media. After the first culture was negative, the second sampling was obtained in the following 72 hours and the final culture sampling was done after 10 days of obtaining negative results from the second culture. In each step, the positive cultures resulted in colonies. Subsequently, diagnostic and biochemical tests were done, and at last, the type of bacteria was characterized. In the current study, all samplings were evaluated for blood culture. All positive blood cultures in the laboratories were assessed and were assayed for antibiotic disc procedure and agar diffusion test with the Kirby and Bauer approach. A suspension of standard separated colonies McFarland formed first and an aseptic swab from bacterial preparation were supplied. Then it was performed in Mueller–Hinton culture medium agar culture in the plate, and antibiotic discs were set with aseptic technique on the plates. Plates were incubated for 24 hours at 37°C and then growth zone was evaluated in comparison with the standard table in these categories: susceptible, resistant, or intermediate. The researchers used antibiotics as reported in their previous study.2 This study was approved by Guilan University of Medical Sciences ethical committee, and all patients filled the written informed consent to participate.
Statistical Analysis
The SPSS software version 16.0 was used for data analysis in all steps. Quantitative data were indicated as mean and standard deviation and qualitative variables as frequency and percentages.
Results
In the period of study, 3,283 blood cultures were sent to these four hospitals’ laboratories. Overall, 2,036 cases (62.01%) were male and others were female. The prevalence of positive cultures was 6.85% (225 cases). In positive cases, the mean age was 46.21±12.49 years old with male preference (male/female: 116/109). The frequency of bacteria isolated from blood cultures of patients is shown in Table 1. Most samples were from Razi Hospital (1,752 cases with 175 positive cases), following Poorsina Hospital (940 cases with five positive cases), Heshmat Hospital (456 cases with 35 positive cases), and Velayat Hospital (136 cases with 10 positive cases).
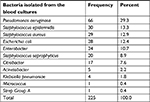  | Table 1 Distribution of bacteria in positive blood culture cases |
The prevalence of bacteria isolated from blood cultures was as follows: Pseudomonas aeruginosa (66, 29.3%), coagulase-negative Staphylococcus (including Staphylococcus epidermidis and Staphylococcus saprophyticus; 50, 22.2%), coagulase-positive Staphylococcus aureus (29, 12.9%), and Escherichia coli (28, 12.4%). These groups included 76.8% of positive cases. A significant relationship was found between gender and the prevalence of bacteria (P=0.031) so that the amount was more common in females. Table 2 shows the usage of antibiotic discs in antibiograms and their patterns.
  | Table 2 Usage of antibiotic discs in antibiograms and their patterns |
According to Table 2, ampicillin, ceftizoxime, ceftriaxone, cotrimoxazole, cefalotin, cefazolin, cefotaxime, ceftazidime, gentamicin, and nitrofurantoin were the top 10 resistant antibiotics.
The pattern of antibiotic resistance to the common bacteria in blood cultures is shown in Table 3.
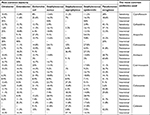  | Table 3 Antibiotic resistance in common bacterial groups in blood cultures performed |
Sensitivity and resistance to used antibiotics are shown in Table 3. As indicated in the table, the most sensitive antibiotics were ciprofloxacin, amikacin, gentamicin, imipenem, nalidixic acid, ceftizoxime, vancomycin, ceftazidime, ceftriaxone, and cotrimoxazole. Most resistance is also related to ampicillin, ceftizoxime, ceftriaxone, cotrimoxazole, cefalotin, cefazolin, cefotaxime, ceftazidime, gentamicin, and nitrofurantoin. When we considered “intermediate results” as “resistance,” the prevalence of resistance in total changed to the top list of resistant antibiotics as shown in Table 4.
  | Table 4 Antibiotic resistance in common bacterial groups in blood cultures performed after adjustment |
Discussion
Antibiotic resistance is a very important issue and despite its critical status, it is growing dangerously. Therefore, investigation of patterns of antibiotic resistance is very important, especially in developing countries, where there is no regular structure for its usage and guideline.5 The current study revealed the bacterial antibiotic resistance and sensitivity pattern in suspicious patients for sepsis workup at four educational referral hospitals of Rasht, Northern Iran.
The rate of positive blood culture in the current observational study was 6.85% (225/3,283) over 1 year. This rate was reported in other studies in developing countries as 13.1% (41/313),10 27.9% (508/1,824),11 59% (215/364),12 18.9% (232/2,130),13 5.17% (456/8,818),14 and 22.9% (43/466).2 Lower rate of positive blood culture in the current study can be due to better education in suitable antibiotic prescription and better management because of educational centers. However, it can be the result of previous antibiotic usage by patients before hospitalization, which masks the rate of positive blood culture. The rate of blood isolate for bacteremia is variable according to geographical region features, and it was reported in India as 16%–44%15,16 and in Iran as 4%–18%.17–19 The most prevalent organisms in the current study were P. aeruginosa, coagulase-negative Staphylococcus, and E. coli. The pattern of every region is unique, however, considering the author’s previous study in the same area,2 it can be suggested that resistance to P. aeruginosa and E. coli was significantly increased after 3 years. This can be related to 1) blind prescription of intravenous antibiotics without indication, 2) lack of attention to guidelines, 3) nonexistence of regional resistant pattern information in hospitals except for research purposes, 4) widespread use of over-the-counter use of antibiotics and easily access for people, and 5) prescriptions by nonhealthcare system-relevant persons at drugstores. Shahidi et al,18 during 3 years, showed that S. aureus, Enterobacter, and P. aeruginosa were the most common bacteria in blood culture samples. Mamishi et al20 reported that the most common bacteria were coagulase-negative Staphylococcus, S. aureus, Klebsiella, E. coli, and P. aeruginosa, respectively. In other studies from Iran, this pattern with some degree of differences was seen; Sedighian et al21: coagulase-negative Staphylococcus, E. coli, P. aeruginosa, and Klebsiella; Kholoujini et al13 and Saderi et al17 reported almost similar pattern for bacteria in blood stream infection.
The most resistant antibiotics in the current study were ampicillin, ceftriaxone, and ceftizoxime. Importantly, these three types of antibiotics are very commonly used at Iranian hospitals intravenously in daily work for management in empirical therapy of sepsis workup and septicemia. The most sensitive antibiotics were ciprofloxacin, gentamicin, and ceftizoxime. In an investigation, maximum resistance to bacteria was 75% and 100% toward ampicillin and tetracycline, respectively.18 In another study, maximum resistance was reported in ampicillin, kanamycin, ceftriaxone, and ceftazidime.22 Accurate diagnosis and efficient therapy are necessary to prevent adverse events of septicemia and reduce the mortality rate from sepsis. Therefore, knowledge of common bacteriological agent and their resistance pattern for prescription of empiric treatment are necessary.
Although there were some similarity in the schema of resistance to antibiotics in comparison with other investigations, the current study showed a unique and specific model in Rasht, which is according to the requirements of health status in Guilan province. Empirical therapy and sometimes unsuitable antibiotics and use of insufficient or higher doses are some causes of resistance to different bacteria. Greater attention should be paid to the findings of antibiograms of samples isolated from blood culture, prevention of over-the-counter antibiotics in drugstores, and free access to people. Some of the limitations were the retrospective nature of the current investigation and lack of control in confounding variables selected for study analysis. Performing large-scale investigations and permanent and prospective evaluation in Guilan province are some necessary strategies to elevate the transition of local knowledge in laboratories to clinical environment for healthcare workers. This can help determine the pattern of antibiotics for the management of empirical and final treatments. It is suggested to utilize resistance pattern and susceptibility in reference laboratories to be more comprehensive. In addition, making registry and provincial network between all laboratories, including public and private in the local environment, can be informative for healthcare workers.
Conclusion
Ceftriaxone was one of the most resistant antibiotic, which could be due to not being used in line with certain guidelines of antibiotic therapy and it is highly suggested to do workshops to use it in its indication. According to significant changes of microorganisms that cause sepsis and meaningful alterations of antibiogram results, the results of this investigation showed that choosing the best treatment for empiric choice should be corrected. The rate of positive samples in the current study was higher than the normal range. Consulting with a pharmacist for antibiotic prescription in hospitals and participation of physicians in antibiotic administration workshops can help decrease resistance.
Disclosure
The authors report no conflicts of interest in this work.
References
Alagesan M, Gopalakrishnan R, Panchatcharam SN, Dorairajan S, Mandayam Ananth T, Venkatasubramanian R. A decade of change in susceptibility patterns of Gram-negative blood culture isolates: a single center study. Germs. 2015;5(3):65–77. | ||
Saeidynia F, Keihanian F, Saeidynia A. Antibiotic resistance in blood culture samples from patients referred to Razi laboratory of Rasht, 2006–2011. Adv Infect Dis. 2014;04(03):165–172. | ||
Saeidinia A, Keihanian F, Delavar SF, Keihanian F, Ranjbar A, Karkan MF. Lack of antibacterial activity of Ruta graveolens extracts against Enterococcus fecalis. Pak J Pharm Sci. 2016;29(4 Suppl):1371–1374. | ||
Gopalakrishnan R, Sureshkumar D. Changing trends in antimicrobial susceptibility and hospital acquired infections over an 8 year period in a tertiary care hospital in relation to introduction of an infection control programme. J Assoc Physicians India. 2010;58(Suppl):25–31. | ||
Jorak A, Keihanian F, Saeidinia A, Heidarzadeh A, Saeidinia F. A cross sectional study on knowledge, attitude and practice of medical students toward antibiotic resistance and its prescription, Iran. Adv Environ Biol. 2014;8(17):675–681. | ||
Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–318. | ||
Zhang X, Gu B, Mei Y, Wen Y, Xia W. Increasing resistance rate to carbapenem among blood culture isolates of Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa in a university-affiliated hospital in China, 2004-2011. J Antibiot. 2015;68(2):115–120. | ||
Saeidinia A, Tavakoli I, Naghipour MR, et al. Prevalence of Strongyloides stercoralis and other intestinal parasites among institutionalized mentally disabled individuals in Rasht, Northern Iran. Iran J Parasitol. 2016;11(4):527–533. | ||
Revelas A. Healthcare - associated infections: a public health problem. Niger Med J. 2012;53(2):59. | ||
Nwadioha I, Odimayo MS, Omotayo J, Olu Taiwo A, Olabiyi E, Taiwo AO. A retrospective cross sectional study of blood culture results in a tertiary hospital, Ekiti, Nigeria. Open J Med Microbiol. 2015;05(04):202–208. | ||
Latif S, Anwar MS, Ahmad I. Bacterial pathogens responsible for blood stream infection (BSI) and pattern of drug resistance in a tertiary care hospital of Lahore. Biomedica. 2009;25(2):101–105. | ||
Chaudhry I, Chaudhry N, Muhammad M, Raheela H, Muhammad T. Etiological pattern of septicemia at three hospitals in Lahore. JCPSP. 2000;10(10):375–379. | ||
Kholoujini M, Karami P, Khaledi A, Neshani A, Matin P, Alikhani MY. Identification of pathogenic bacteria in blood cultures and susceptibility testing of isolates with various antibiotics. Avicenna J Clin Microb Infec. 2016;3(3):e37480. | ||
Barati M, Taher MT, Abasi R, Zadeh MM, Barati M, Shamshiri AR. Bacteriological profile and antimicrobial. Arch Clin Infect Dis. 2009;4(2):87–95. | ||
Khanal B, Harish BN, Sethuraman KR, Srinivasan S. Infective endocarditis: report of a prospective study in an Indian hospital. Trop Doct. 2002;32(2):83–85. | ||
Sharma M, Goel N, Chaudhary U, Aggarwal R, Arora DR. Bacteraemia in children. Indian J Pediatr. 2002;69(12):1029–1032. | ||
Saderi H, Karimi A, Looni M. The frequency of bacteria isolated from blood cultures and their antibiotic susceptibility patterns in a university hospital in Tehran [In Persian]. Iran South Med J. 2009;12(2):142–148. | ||
Shahidi KMD, Amir G, Ayravani M. Studying the blood cultures of admitted patients for the common microorganisms, epidemiological changes and antibiogram in Amir hospital from 1377-79 [In Persian]. Tehran Univ Med J. 2002;60(1):7–31. | ||
Mohajeri P, Izadi B, Naghshi N. Antibiotic sensitivity of Escherichia coli isolated from urinary tract infection referred to Kermanshah central laboratory. Behbood. 2011;15(1):51–56. | ||
Mamishi S, Pourakbari B, Ashtiani MH, Hashemi FB. Frequency of isolation and antimicrobial susceptibility of bacteria isolated from bloodstream infections at Children’s Medical Center, Tehran, Iran, 1996-2000. Int J Antimicrob Agents. 2005;26(5):373–379. | ||
Sedighian F, Sanee A, Alaouddoulee H, Arshi M, Rekabpoor K. The Study of antibiotic resistance of microorganisms isolated in Yahya nejad Hospital, Babol (North of Iran), 1385. Med Lab J. 2008;2(2):29–35. | ||
Habibian R, Imani R, Khoshdel A. Changes of trend of antibiotic susceptibility in isolated bacteria from culture of ICU patients of Shahrekord Ayatollah Kashanani Hospital, I. R. Iran. J Shahrekord Univ Med Sci. 2012;14(1):77–83. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
