Back to Journals » Infection and Drug Resistance » Volume 15
Application of Droplet Digital PCR to Detection of Mycobacterium tuberculosis and Mycobacterium leprae Infections: A Narrative Review
Authors Fan Y, Chen J, Liu M, Xu X, Zhang Y, Yue P, Cao W, Ji Z, Su X, Wen S, Kong J, Zhou G, Li B, Dong Y, Liu A, Bao F
Received 22 November 2021
Accepted for publication 25 January 2022
Published 15 March 2022 Volume 2022:15 Pages 1067—1076
DOI https://doi.org/10.2147/IDR.S349607
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Yuxin Fan,1– 3 Jingjing Chen,1,4 Meixiao Liu,1,2 Xin Xu,1,2 Yu Zhang,1– 3 Peng Yue,1,4 Wenjing Cao,1,4 Zhenhua Ji,1,2 Xuan Su,1,2 Shiyuan Wen,1– 3 Jing Kong,1,2 Guozhong Zhou,1,2 Bingxue Li,1,2 Yan Dong,1,2 Aihua Liu,1,3,4 Fukai Bao1– 3
1The Institute for Tropical Medicine, Faculty of Basic Medical Sciences, Kunming Medical University, Kunming, 650500, People’s Republic of China; 2Department of Microbiology and Immunology, Kunming Medical University, Kunming, 650030, People’s Republic of China; 3Yunnan Province Key Laboratory of Children’s Major Diseases Research, The Affiliated Children Hospital, Kunming Medical University, Kunming, 650030, People’s Republic of China; 4Department of Biochemistry and Molecular Biology, Kunming Medical University, Kunming, 650030, People’s Republic of China
Correspondence: Aihua Liu; Fukai Bao, The Institute for Tropical Medicine, Faculty of Basic Medical Sciences, Kunming Medical University, Kunming, 650500, People’s Republic of China, Email [email protected]; [email protected]
Abstract: Tuberculosis (TB) is a chronic infectious disease caused by Mycobacterium tuberculosis (MTB) infection, which has seriously endangered human health for many years. With the emergence of multidrug-resistant and extensively drug-resistant MTB, the prevention and treatment of TB has become a pressing need. Early diagnosis, drug resistance monitoring, and control of disease transmission are critical aspects in the prevention and treatment of TB. However, the currently available diagnostic technologies and drug sensitivity tests are time consuming, and thus, it is difficult to achieve the goal of early diagnosis and detection drug sensitivity, which results in limited control of disease transmission. The development of molecular testing technology has gradually achieved the vision of rapid and accurate diagnosis of TB. Droplet digital PCR (ddPCR) is an excellent nucleic acid quantification method with high sensitivity and no need for a calibration curve. Herein, we review the application of ddPCR in TB diagnosis and drug resistance detection and transmission monitoring.
Keywords: Mycobacterium tuberculosis, tuberculosis, droplet digital PCR, diagnosis, drug resistance test, disease transmission
Summary
Early diagnosis, drug resistance monitoring, and control of disease transmission are critical aspects in the prevention and treatment of TB. However, the currently available diagnostic technologies and drug sensitivity tests are time consuming, and thus, difficult to achieve the goal of early diagnosis and detection drug sensitivity. Droplet digital PCR (ddPCR) is an excellent nucleic acid quantification method with high sensitivity and its application in TB diagnosis and drug resistance detection and transmission monitoring is worth exploring.
Introduction
Tuberculosis (TB) is caused by Mycobacterium tuberculosis (MTB) infection, a disease which poses a serious threat to human health and kills nearly two million individuals each year.1 The World Health Organization (WHO) and the 194 member states of the United Nations approved the WHO strategy to end TB at the 2014 World Health Assembly, aiming to reduce the number of TB deaths by 95% and to lower the new incidence rate of TB diagnoses by 90% between 2015 and 2035.2 According to the WHO Global TB Report 2020, there were nearly 10 million new TB patients and 2 billion latent TB patients around the world in 2019.1 Despite the global decline in TB incidence, it is not sufficient to meet the 2020 milestone target of the End TB Strategy (a 20% reduction in TB incidence by 2015–2020).1 TB control remains critical.
The rapid development of molecular biology technology has overcome the limitations of time-consuming and low sensitivity of acid-fast staining and culture for TB diagnosis, so a rapid and accurate diagnosis of TB can gradually be achieved. Molecular detection of persistent genes also significantly reduces the time required for drug sensitivity testing, which typically requires two to three months to complete phenotypic drug sensitivity assays. Innovative diagnostic tools with greater specificity, sensitivity, and automation are being developed. This paper introduces a new molecular diagnostic technique, droplet digital PCR (ddPCR), and its application in TB, with the hope of stimulating new ideas for the prevention and treatment of TB.
Droplet Digital Polymerase Chain Reaction Technology
Over time, continuous and in-depth development of molecular biology approaches have paved the way for the diagnosis of early and latent infection of pathogens. Real-time quantitative polymerase chain reaction (qPCR) has been used as a routine diagnostic tool for gene expression and quantitative determination of deoxyribonucleic acid transcripts.3 qPCR is based on PCR, and fluorescence reading represents the amount of fluorescence signal after each cycle, which allows to quantify the target relative to the calibration substance.4,5 Although qPCR remains the gold standard for nucleic acid quantification, it is not suitable for the detection of small differences.6 Furthermore, the quantification of qPCR is generally inaccurate, as it depends on comparing unknown samples with a standard curve.3 The ddPCR approach based on digital polymerase chain reaction (dPCR) is a new absolute nucleic acid quantification method. It combines microfluidic technology with the PCR, allowing accurate quantification of a single copy of DNA and achieving accurate quantification of target DNA with high sensitivity and specificity.7,8 ddPCR can separate samples into thousands of drops and performs independent PCR sub-reactions so that each sub-reaction contains little or no off-target sequence.5 After the PCR reaction, the fluorescence signal is detected in each droplet. Poisson statistical analysis is performed on the proportion of positive droplets to achieve an accurate quantification of the target sequence (Figure 1).3,9
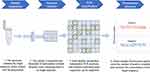 |
Figure 1 Principle and workflow of ddPCR. |
Conventional qPCR amplifies all target molecules in the sample, and the signal obtained will represent the average signal of the different DNA sequences present in the sample. ddPCR can amplify each target gene in a separate compartment, making detection more specific and sensitive.10 Compared to qPCR, ddPCR can quantify DNA without needing a standard curve. Furthermore, the quantification of ddPCR is based on binomial statistics and its inherent accuracy and performance indicators are defined mathematically.11–13 The small volume of the reaction not only greatly improves the sensitivity of the reaction, but also reduces template competition, giving ddPCR greater resistance to various inhibitors.5 These characteristics make ddPCR an accurate target DNA quantitative method with high sensitivity and specificity and an ideal alternative method to detect infectious agents with extremely low-level load such as in early stage infections and latent infections.12 Currently, several microfluidic platforms based on ddPCR are being commercialized, including RaindropTM digital PCR (Raindance Technologies), Bio-Rad QX200TM droplet digital system (Bio-Rad Laboratories), and the NaicaTM system (Stilla Technologies).14
Application of ddPCR to Diagnosis of Mycobacterium tuberculosis Infection
Rapid diagnosis of TB is very important for patient treatment and infection control. TB is usually diagnosed using qualitative methods, such as smear acid-fast staining method, specimen culture, which are the most common diagnostic methods for TB, and clinical decisions depend on the presence or absence of the pathogen15,16. However, the culture of Mycobacterium tuberculosis is time-consuming and the sensitivity of acid-fast bacilli (AFB) smear is low.17 The development of molecular methods provides the possibility for a simple, rapid, and objective diagnosis of TB. ddPCR is an emerging technology that enables absolute nucleic acid quantification without the use of a standard curve.12 To date, an increasing number of studies have indicated that ddPCR is a promising tool for diagnosing infectious diseases in low-copy samples.18 In 2015, Devonshire et al19 used the MTB DNA template to evaluate the effects of template type, reaction mixture on the performance of ddPCR. They found that ddPCR was repeatable quantitative DNA method. In their subsequent study,16 they used ddPCR to quantify MTB in artificial sputum and found that ddPCR also had satisfactory repeatability and accuracy for the quantification of MTB. The MTB-specific sequence detection method based on ddPCR is expected to develop into a minimally invasive, rapid, and accurate diagnostic choice.
Quantification of Mycobacterium tuberculosis
Pathogen quantification is a potential indicator for determining treatment outcomes and predicting recurrence, and quantification is increasingly proposed as a tool to help manage TB infections. The application of ddPCR provides a new method for the accurate quantification of MTB pathogens. In 2016, Ushio et al15 successfully detected and quantified MTB in plasma of TB patients by ddPCR for the first time. ddPCR can also be used to quantify low-copy target molecules with higher accuracy and sensitivity. In 2017, our research group20 used ddPCR to detect MTB-specific DNA targets (IS6110) in whole blood samples. The results show that ddPCR can be used to detect MTB-specific sequences from whole blood. This is the first time ddPCR was used in the clinical diagnosis of TB, providing a basis for the exploration of ddPCR as a quantitative molecular diagnostic test for infectious diseases. Subsequently, Song et al21 applied ddPCR to the diagnosis of infantile TB patients. They found that ddPCR also showed advantages of absolute quantification and high sensitivity in whole blood specimens from infants who lacked early respiratory symptoms and from whom it was difficult to obtain sputum.
Diagnosis of Tuberculosis in Patients with HIV Infection
In 2018, Yamamoto et al22 used ddPCR to detect MTB in the plasma of a patient with severe immune deficiency. Neither acid-fast staining nor qPCR of sputum, blood, and urine samples from this patient detected the presence of MTB, while the ddPCR results showed positive amplification of MTB-specific sequences (IS6110 and gyrB). The presence of disseminated MTB infection was confirmed in the patient at a later autopsy. MTB co-infection with human immunodeficiency virus (HIV) accounts for 8.2% of new cases of TB and is the most common cause of death from TB.1 For various reasons, patient sputum samples may not be available or sputum samples are negative, which result in an ineffective and timely diagnosis of TB. Although the detection of lipoarabinomannan (LAM) in urine has been applied in the diagnosis of TB in HIV patients, existing commercial kits still lack sufficient sensitivity.22
Mycobacterium tuberculosis Detection in Different Samples
After Yang et al20 demonstrated, ddPCR could be used to detect MTB in whole blood samples, studies on the use of ddPCR for a series of studies investigating the detection of MTB in pathological samples, exosomes, and other specimens have been developed. In 2020, Cho et al17 provided a new method to detect the MTB specific sequence in exosomes (IS6110) by ddPCR. They used ddPCR to detect 190 respiratory tract samples from suspected TB patients. Compared to culture, the ddPCR showed better sensitivity and specificity, and the ddPCR assay using exoDNA had higher sensitivity than total DNA.
In the same year, Cao et al23 used ddPCR to detect MTB specific sequences (IS6110) in 65 pathological samples and evaluated the sensitivity of ddPCR to detect MTB in formalin-fixed and paraffin-embedded (FFPE) specimens. Of 65 samples, 45 were identified as probable TB patients (Ct value was between 37 and 40) using qPCR. ddPCR improved the positive rate of probable TB patients by 57.8%, which showed a higher sensitivity of ddPCR for the detection of MTB in FFPE samples.23
The insertion sequence (IS) has been used for the diagnosis of MTB, the most commonly used of which is IS6100,24 but some difficulties remain. On the one hand, IS6110-based diagnosis has been proven to be hampered by low copy numbers or IS6110 repeat deletions.25 The ddPCR with highly sensitivity can be a good way to alleviate this problem. On the other hand, some clinical MTB strains show IS6110 negativity, which can lead to false negative results. In this regard, Nyaruaba et al26 developed and evaluated a single dye duplex ddPCR detection method, which can reliably quantify two MTB targets in the same channel. They used the single dye double-stranded ddPCR method combined with two genes IS6100 and IS1081 for the quantification of TB. This double-target joint detection method helps to eliminate false negative results, and the single-dye duplex technology also greatly reduces the cost of detection, laying the foundation for the popularization of ddPCR.
Detection of Latent Tuberculosis Infection
In a low prevalence environment, most active diseases (47–87%) are due to the reactivation of latent TB infection (LTBI).27 However, there is no gold standard for the diagnosis of latent TB infection, which means that some TB patients do not have access to appropriate treatment.28 The tuberculin skin test and the interferon-γ release test (IGRA) also have limitations in the diagnosis of LTBI, and it is difficult to determine whether the latent infection of MTB would develop into active TB.29,30
Harboring of MTB in CD34 + peripheral blood mononuclear cells overcomes adverse conditions, including hypoxia, immune attack, and drug treatment, thus allowing long-term presence in the host.27 Hematopoietic stem cells carrying CD34 glycoprotein as a surface marker may represent ecological niches of MTB during latent TB infection, and detection of the presence of MTB in these cells may provide valuable information for the diagnosis of latent TB.27 In 2021, Belay et al31 used ddPCR to detect MTB in peripheral blood mononuclear CD34 + cells from asymptomatic adults testing positive for the IGRA. This indicates that the detection of MTB DNA in peripheral blood mononuclear cells has potential applications in the diagnosis, monitoring, and preventive treatment of latent TB infection. However, this approach requires the isolation of individual nucleated cells, which is cumbersome and limits its clinical application.
Detection of Mycobacterium leprae Infection
The application of ddPCR in the diagnosis of Mycobacterium leprae infection has also been studied. In 2019, Cheng et al18 developed a ddPCR detection method for the diagnosis of leprosy. They used qPCR and ddPCR to detect two sensitive DNA targets (RLEP and GroEL) of leprosy bacilli and then evaluated the sensitivity and specificity of the method. They found a higher sensitivity of ddPCR compared to qPCR for the detection of Mycobacterium leprae in skin biopsy specimens with paucibacillary (PB) load from patients (79.5% vs 36.4%).
Application of ddPCR in the Analysis of MTB Drug Resistance
Multidrug-resistant TB (MDR-TB) remains a major public health concern in many countries, with data published by WHO 2020 showing that the average global success rate of MDR-TB treatment is only 57%.1 Increasing incidence of MDR and extensively drug-resistant TB cases due to non-compliance with drug regimens, misuse, or misadministration, and a cumulative reduction in global TB incidence of only 9% from 2015–2019.32 The gold standard drug susceptibility test (DST) is based on culture, using an agar-based media indirect proportion method. But it is time-consuming and the reliability of the results varies depending on the drug test, the bacterial concentration, the culture medium, and the culture time.33,34 Furthermore, the effects of heterologous resistant bacteria on DST remains unclear.35 Some TB patients present a combination of drug-sensitive and drug-resistant organisms, a phenomenon known as heteroresistance.36,37 This is common in MTB and is believed to lead to worse treatment outcomes in TB.38,39 To achieve early identification of drug resistance for effective treatment and clinical management of TB, it is necessary to improve the speed and specificity of DST.
The main reason for drug resistance in clinical MTB is chromosomal mutations, thus, genotyping can quickly identify the main drug resistance mechanism, such as rpob mutation inducing rifampicin resistance.34,40 Methods based on conventional PCR, such as Xpert MTB/RIF, linear probes, and sequencing, can rapidly detect MTB bacteria and their sensitivity to first-line and second-line drugs without the need for culture.33,41 Genotypic approaches to screening of resistance hotspot mutations have also been found to be easier to implement in low-income countries, as they avoid culture and biosafety constraints.34 However, the low number of mutation templates detected by conventional PCR is limited because rich sequences may be amplified preferentially, and a relative proportion of 10% of drug-resistant organisms are required for linear probe detection, or Xpert MTB/RIF detection, 65–100% of the mutated RPOB DNA is required.36,42,43 The ddPCR not only has the advantage of high sensitivity without the need for a calibration curve for the absolute quantification of nucleic acids but it also presents incomparable precision in the detection of copy number variation (CNV), in which it can achieve smaller variations in measurement.44 By modifying the ddPCR program, such as primers, probes, and amplification conditions, ddPCR can accurately detect rare mutations in wild-type sequences, including resistant subgroups.36,38,45 Whale et al44 evaluated the potential accuracy of dPCR for detecting mutations and found that dPCR could detect much smaller CNVs than qPCR in the same experimental replication. Subsequently, Pholwat et al36 also explored the role of dPCR in the detection and quantification of heterologous resistance in mixed TB populations. The dPCR method was able to identify mutant sequences in mixtures containing 1000:1 susceptible: drug-resistant TB.36
In 2015, Taylor et al45 used optimized ddPCR to detect oseltamivir-resistant influenza mutations (H275Y) in influenza patients, and the results were accurate, interpretable, and statistically significant, with a 30-fold increase in sensitivity compared to qPCR. In 2019, Rigouts et al38 also used ddPCR as a reference method to determine the limit of detection (LOD) of MTB gatifloxacin resistant mutants in the genotype test. In the same year, Luo et al33 established a culture- and ddPCR-based drug sensitivity test method for MTB. The MTB target sequence (IS6110) was detected by ddPCR. If the results of ddPCR were positive, the bacterial solution was inoculated into 7H9 medium for the drug sensitivity test. Finally, the DNA fold change of the samples at 0 d and 4 d after culture were determined by ddPCR, and the DST results were calculated. The results showed that culture ddPCR allowed rapid detection of MTB and multidrug-resistant TB directly from sputum within 4 days. Compared with other rapid DST methods, culture ddPCR has the advantage of high specificity, fast detection speed, and more comprehensive detection range.33 Because the primers and probes for culture ddPCR are specific to the IS6110 fragment of MTB and have high specificity; the assay is based on measuring changes in the amount of DNA during MTB growth and the time required to measure growth is much shorter.33 Culture ddPCR can also identify all types of drug resistance, including resistance due to unknown mechanisms, providing a greater range of detection than other molecular methods that can only detect hotspot regions.33 In addition to routine examination of drug resistance, ddPCR can be applied to the analysis of drug resistance mechanisms, Singh et al46 studied the mechanism of antibacterial resistance to 5-fluorouracil (5-FU) in TB.
The ddPCR has good precision in detecting CNV, which makes it possible to detect mutations involved in drug resistance quickly and accurately. However, no molecular biology-based resistance detection technology is currently available for new drugs such as bedaquiline and delamanid, and the discovery of new resistance mutation loci can broaden the application area of ddPCR.47 The combination of highly sensitive ddPCR and the culture method can greatly shorten the detection time of drug resistance and provide a great prospect for the early detection of drug resistance in TB.
Use of ddPCR in the Hospital Setting to Measure Infectivity
Early diagnosis and drug sensitivity testing are important aspects of anti-TB treatment. Despite the significant efforts, the rate of reduction in TB incidence is still insufficient and treatment alone is unlikely to significantly reduce the disease burden. It is very important to control the spread of TB for the prevention and treatment of TB. The commonly used approach is through the epidemiological survey of contact tracing. But contact tracking is difficult to implement in resource-limited environments, and large-scale epidemiological studies or clinical applications are too cumbersome, so new approaches are urgently needed to assess the spread of TB to control TB.48,49
MTB is spread by aerosols from TB patients (5-mm infectious air droplets).50 By collecting air samples from suspected environments and populations and testing them, it may serve as a reliable method to assess the spread of MTB. In a retrospective study of patients with TB, cough aerosols were found to be a more specific specimen to assess infectivity than sputum smears, although they were of little diagnostic use.48 Identification of disease-causing particles has traditionally relied on microscopy, media culture, or immunoassay methods.51 However, in the analysis of MTB in air, due to the low concentration of naturally occurring MTB particles and the dilution characteristics of airborne microorganisms, the use of culture is susceptible to contamination by other bacteria and fungi.52,53 Furthermore, MTB growth is slow and usually takes 28 days to culture on Löwenstein-Jensen medium, which does not meet the rapid detection requirement.54 Fortunately, with the development of molecular techniques, the combination of air sampling and molecular diagnostics has advanced the study of MTB in air-borne transmission. Mastorides et al55 performed an air sampling analysis of MTB using a combination of air filtration and PCR. Subsequently, several studies have used air sampling with qPCR to assess airborne MTB in the health care settings, including several studies in Taipei, Slovenia, South Africa, and Thailand, all of which reported good results, suggesting that air sampling combined with qPCR is a feasible method to assess the risk of airborne TB exposure.56–59 Patterson et al52 used a microbial culture method and ddPCR to detect MTB in bioaerosols. In the study, environmental bioaerosol from 35 newly diagnosed but untreated TB patients tested positive for MTB culture in 42.8% and for ddPCR in 92.96%.52 The combination of high-sensitivity air sampling and detection technology offers unlimited possibilities for the detection of MTB, although the molecular approach still has limitations.59 PCR signals cannot distinguish between living and dead bacteria, so detection represents only the estimated risk of TB transmission, not the actual risk. Routine surveillance of airborne MTB is critical to the safety of health care workers together with the interruption of TB transmission. Airborne sample collection and highly sensitive molecular diagnostic methods offer new opportunities in this effort. Although the application of ddPCR in airborne MTB has been less studied, it has unlimited potential, and additional research is needed before the effective control of TB transmission can be achieved. Summary of studies exploring the use of ddPCR in Mycobacterium infection is also provided in Table 1.
 |
Table 1 Summary of Studies Exploring the Use of ddPCR in Mycobacterium tuberculosis and Mycobacterium leprae Infections |
Conclusions
ddPCR is a promising novel detection technique that can be used for CNV analysis, rare mutation detection, single nucleotide polymorphism genotyping, and transcription quantification.7,13 The combination of microfluidic technology and PCR gives ddPCR unique characteristics and is increasingly used in clinical fields such as infectious diseases, tumors, virus copy number analysis.10,60 Herein, we reviewed the application of ddPCR in TB diagnosis, drug resistance analysis, and aerosol transmission. The ddPCR is a quantitative method for the detection of nucleic acid with high sensitivity, high reproducibility, and no calibration curve. Using ddPCR to detect MTB-specific sequences in samples may become a new method for TB diagnosis. The ddPCR detection of drug resistance mutations to MTB culture greatly shortens the time of drug sensitivity testing, provides timely and accurate information to guide clinical use, and is expected to reduce the global burden of widespread drug resistance and the gradual increase of MDR.
However, there are still several limitations that need to be overcome in order to widely use ddPCR in clinical settings. Firstly, although instruments and reagents for ddPCR are commercially available, their high price limits their widespread use. Secondly, the selection of detection targets and the setting of detection thresholds are also challenging for the practical application of ddPCR in clinical practice. The setting of the threshold lines for positive and negative droplet may bias the results. To make ddPCR more useful in MTB detection, a suitable detection threshold must be set. In addition, the significance of ddPCR detection of trace DNA in the diagnosis, treatment and prevention of clinical diseases should be further explored. In conclusion, cost reduction and standardization of ddPCR procedures and conditions are necessary to make ddPCR widely available for clinical use.
Despite some limitations, ddPCR has great potential in controlling airborne transmission of TB, greater efforts are warranted to explore this method for containing MTB and protecting human health.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China and Yunnan Province Department of Science and Technology-Kunming Medical University Joint Fund Projects. The funding institutions had no involvement in the design of the study or review of the manuscript.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
National Natural Science Foundation of China (No.82160304, 81560596, 81860644) and Joint Foundation of Yunnan Province Department of Science, Technology-Kunming Medical University [No. 2019FE001 (−002) and 2017FE467 (−001)].
Disclosure
The authors report no conflicts of interest in this work.
References
1. World Health Organization. Global Tuberculosis Report 2020 [M]. Geneva: World Health Organization; 2020.
2. Uplekar M, Weil D, Lonnroth K, et al. WHO’s new end TB strategy. Lancet. 2015;385(9979):1799–1801. doi:10.1016/S0140-6736(15)60570-0
3. Vossen RH, White SJ. Quantitative DNA analysis using droplet digital PCR. Methods Mol Biol. 2017;1492:167–177.
4. Hawkins SFC, Guest PC. Multiplex analyses using real-time quantitative PCR. Methods Mol Biol. 2017;1546:125–133.
5. Quan PL, Sauzade M, Brouzes E. dPCR: a technology review. Sensors. 2018;18(4):1271. doi:10.3390/s18041271
6. Taylor SC, Nadeau K, Abbasi M, Lachance C, Nguyen M, Fenrich J. The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37(7):761–774. doi:10.1016/j.tibtech.2018.12.002
7. Mazaika E, Homsy J. Digital droplet PCR: CNV analysis and other applications. Curr Protoc Hum Genet. 2014;82:
8. Parkin B. Rare variant quantitation using droplet digital PCR. Methods Mol Biol. 2019;1881:239–251.
9. Whale AS, Cowen S, Foy CA, Huggett JF. Methods for applying accurate digital PCR analysis on low copy DNA samples. PLoS One. 2013;8(3):e58177. doi:10.1371/journal.pone.0058177
10. Perkins G, Lu H, Garlan F, Taly V. Droplet-based digital PCR: application in cancer research. Adv Clin Chem. 2017;79:43–91.
11. Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 2015;61(1):79–88. doi:10.1373/clinchem.2014.221366
12. Colafigli G, Scalzulli E, Di Prima A, et al. Digital droplet PCR as a predictive tool for successful discontinuation outcome in chronic myeloid leukemia: is it time to introduce it in the clinical practice? Crit Rev Oncol Hematol. 2021;157:103163. doi:10.1016/j.critrevonc.2020.103163
13. Nyaruaba R, Mwaliko C, Kering KK, Wei H. Droplet digital PCR applications in the tuberculosis world. Tuberculosis. 2019;117:85–92. doi:10.1016/j.tube.2019.07.001
14. Postel M, Roosen A, Laurent-Puig P, Taly V, Wang-Renault SF. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Rev Mol Diagn. 2018;18(1):7–17. doi:10.1080/14737159.2018.1400384
15. Ushio R, Yamamoto M, Nakashima K, et al. Digital PCR assay detection of circulating Mycobacterium tuberculosis DNA in pulmonary tuberculosis patient plasma. Tuberculosis. 2016;99:47–53. doi:10.1016/j.tube.2016.04.004
16. Devonshire AS, O’Sullivan DM, Honeyborne I, et al. The use of digital PCR to improve the application of quantitative molecular diagnostic methods for tuberculosis. BMC Infect Dis. 2016;16(1):1–10. doi:10.1186/s12879-016-1696-7
17. Cho SM, Shin S, Kim Y, et al. A novel approach for tuberculosis diagnosis using exosomal DNA and droplet digital PCR. Clin Microbiol Infect. 2020;26(7):
18. Cheng X, Sun L, Zhao Q, et al. Development and evaluation of a droplet digital PCR assay for the diagnosis of paucibacillary leprosy in skin biopsy specimens. PLoS Negl Trop Dis. 2019;13(3):e0007284. doi:10.1371/journal.pntd.0007284
19. Devonshire AS, Honeyborne I, Gutteridge A, et al. Highly reproducible absolute quantification of Mycobacterium tuberculosis complex by digital PCR. Anal Chem. 2015;87(7):3706–3713. doi:10.1021/ac5041617
20. Yang J, Han X, Liu A, et al. Use of digital droplet PCR to detect Mycobacterium tuberculosis DNA in whole blood-derived DNA samples from patients with pulmonary and extrapulmonary tuberculosis. Front Cell Infect Microbiol. 2017;7:369. doi:10.3389/fcimb.2017.00369
21. Song N, Tan Y, Zhang L, et al. Detection of circulating Mycobacterium tuberculosis-specific DNA by droplet digital PCR for vaccine evaluation in challenged monkeys and TB diagnosis. Emerg Microbes Infect. 2018;7(1):1–9. doi:10.1038/s41426-017-0002-0
22. Yamamoto M, Ushio R, Watanabe H, et al. Detection of Mycobacterium tuberculosis-derived DNA in circulating cell-free DNA from a patient with disseminated infection using digital PCR. Int J Infect Dis. 2018;66:80–82. doi:10.1016/j.ijid.2017.11.018
23. Cao Z, Wu W, Wei H, et al. Using droplet digital PCR in the detection of Mycobacterium tuberculosis DNA in FFPE samples. Int J Infect Dis. 2020;99:77–83. doi:10.1016/j.ijid.2020.07.045
24. Sankar S, Kuppanan S, Balakrishnan B, Nandagopal B. Analysis of sequence diversity among IS6110 sequence of Mycobacterium tuberculosis: possible implications for PCR based detection. Bioinformation. 2011;6(7):283–285. doi:10.6026/97320630006283
25. McEvoy CR, Falmer AA, Gey van Pittius NC. The role of IS6110 in the evolution of Mycobacterium tuberculosis. Tuberculosis. 2007;87(5):393–404. doi:10.1016/j.tube.2007.05.010
26. Nyaruaba R, Xiong J, Mwaliko C, et al. Development and evaluation of a single dye duplex droplet digital PCR assay for the rapid detection and quantification of Mycobacterium tuberculosis. Microorganisms. 2020;8(5):701. doi:10.3390/microorganisms8050701
27. Mayito J, Andia Biraro I, T. Reece S, R. Martineau A, P. Kateete D. Detection of Mycobacterium tuberculosis DNA in CD34+ peripheral blood mononuclear cells of Ugandan adults with latent infection: a cross-sectional and nested prospective study. AAS Open Res. 2020;3:34. doi:10.12688/aasopenres.13108.1
28. Wang CC, Zhu B, Fan X, Gicquel B, Zhang Y. Systems approach to tuberculosis vaccine development. Respirology. 2013;18(3):412–420. doi:10.1111/resp.12052
29. Huebner RE, Schein MF, Bass JB
30. Barth RE, Mudrikova T, Hoepelman AI. Interferon-gamma release assays (IGRAs) in high-endemic settings: could they play a role in optimizing global TB diagnostics? Evaluating the possibilities of using IGRAs to diagnose active TB in a rural African setting. Int J Infect Dis. 2008;12(6):e1–e6. doi:10.1016/j.ijid.2008.03.026
31. Belay M, Tulu B, Younis S, et al. Detection of Mycobacterium tuberculosis complex DNA in CD34-positive peripheral blood mononuclear cells of asymptomatic tuberculosis contacts: an observational study. Lancet Microbe. 2021;2(6):e267–e275. doi:10.1016/S2666-5247(21)00043-4
32. World Health Organization. Global Tuberculosis Report 2019 [M]. Geneva: World Health Organization; 2019.
33. Luo J, Luo M, Li J, et al. Rapid direct drug susceptibility testing of Mycobacterium tuberculosis based on culture droplet digital polymerase chain reaction. Int J Tuberc Lung Dis. 2019;23(2):219–225. doi:10.5588/ijtld.18.0182
34. Miotto P, Zhang Y, Cirillo DM, Yam WC. Drug resistance mechanisms and drug susceptibility testing for tuberculosis. Respirology. 2018;23(12):1098–1113. doi:10.1111/resp.13393
35. Folkvardsen DB, Svensson E, Thomsen V, et al. Can molecular methods detect 1% isoniazid resistance in Mycobacterium tuberculosis? J Clin Microbiol. 2013;51(5):1596–1599. doi:10.1128/JCM.00472-13
36. Pholwat S, Stroup S, Foongladda S, Houpt E. Digital PCR to detect and quantify heteroresistance in drug resistant Mycobacterium tuberculosis. PLoS One. 2013;8(2):e57238. doi:10.1371/journal.pone.0057238
37. Morand B, Mühlemann K. Heteroresistance to penicillin in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 2007;104(35):14098–14103. 23468945. doi:10.1073/pnas.0702377104
38. Rigouts L, Miotto P, Schats M, et al. Fluoroquinolone heteroresistance in Mycobacterium tuberculosis: detection by genotypic and phenotypic assays in experimentally mixed populations. Sci Rep. 2019;9(1):1–8. doi:10.1038/s41598-019-48289-9
39. Zetola NM, Shin SS, Tumedi KA, et al. Mixed Mycobacterium tuberculosis complex infections and false-negative results for rifampin resistance by GeneXpert MTB/RIF are associated with poor clinical outcomes. J Clin Microbiol. 2014;52(7):2422–2429. 24789181. doi:10.1128/JCM.02489-13
40. Schön T, Miotto P, Köser CU, Viveiros M, Böttger E, Cambau E. Mycobacterium tuberculosis drug-resistance testing: challenges, recent developments and perspectives. Clin Microbiol Infect. 2017;23(3):154–160. 27810467. doi:10.1016/j.cmi.2016.10.022
41. Gardee Y, Dreyer AW, Koornhof HJ, et al. Evaluation of the GenoType MTBDRsl version 2.0 assay for second-line drug resistance detection of Mycobacterium tuberculosis isolates in South Africa. J Clin Microbiol. 2017;55(3):791–800. doi:10.1128/JCM.01865-16
42. Hofmann-Thiel S, van Ingen J, Feldmann K, et al. Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur Respir J. 2009;33(2):368–374. doi:10.1183/09031936.00089808
43. Blakemore R, Story E, Helb D, et al. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J Clin Microbiol. 2010;48(7):2495–2501. doi:10.1128/JCM.00128-10
44. Whale AS, Huggett JF, Cowen S, et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012;40(11):e82. doi:10.1093/nar/gks203
45. Taylor SC, Carbonneau J, Shelton DN, Boivin G. Optimization of Droplet Digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: clinical implications for quantification of Oseltamivir-resistant subpopulations. J Virol Methods. 2015;224:58–66. doi:10.1016/j.jviromet.2015.08.014
46. Singh V, Brecik M, Mukherjee R, et al. The complex mechanism of antimycobacterial action of 5-fluorouracil. Chem Biol. 2015;22(1):63–75. doi:10.1016/j.chembiol.2014.11.006
47. World Health Organization. Update on the Use of Nucleic Acid Amplification Tests to Detect TB and Drug-Resistant TB: Rapid Communication[M]. Geneva: World Health Organization; 2021.
48. Jones-López EC, Acuña-Villaorduña C, Ssebidandi M, et al. Cough aerosols of Mycobacterium tuberculosis in the prediction of incident tuberculosis disease in household contacts. Clin Infect Dis. 2016;63(1):10–20. doi:10.1093/cid/ciw199
49. Zürcher K, Morrow C, Riou J, et al. Novel approach to estimate tuberculosis transmission in primary care clinics in sub-Saharan Africa: protocol of a prospective study. BMJ Open. 2020;10(8):e036214. doi:10.1136/bmjopen-2019-036214
50. Fennelly KP, Jones-López EC, Ayakaka I, et al. Variability of infectious aerosols produced during coughing by patients with pulmonary tuberculosis. Am J Respir Crit Care Med. 2012;186(5):450–457. doi:10.1164/rccm.201203-0444OC
51. West JS, Atkins SD, Emberlin J, Fitt BD. PCR to predict risk of airborne disease. Trends Microbiol. 2008;16(8):380–387. doi:10.1016/j.tim.2008.05.004
52. Patterson B, Morrow C, Singh V, et al. Detection of Mycobacterium tuberculosis bacilli in bio-aerosols from untreated TB patients. Gates Open Res. 2017;1:11. doi:10.12688/gatesopenres.12758.1
53. Roy CJ, Milton DK. Airborne transmission of communicable infection–the elusive pathway. N Engl J Med. 2004;350(17):1710–1712. doi:10.1056/NEJMp048051
54. Jing W, Jiang X, Zhao W, Liu S, Cheng X, Sui G. Microfluidic platform for direct capture and analysis of airborne Mycobacterium tuberculosis. Anal Chem. 2014;86(12):5815–5821. doi:10.1021/ac500578h
55. Mastorides SM, Oehler RL, Greene JN, Sinnott J, Kranik M, Sandin RL. The detection of airborne Mycobacterium tuberculosis using micropore membrane air sampling and polymerase chain reaction. Chest. 1999;115(1):19–25. doi:10.1378/chest.115.1.19
56. Chen P-S, Li C-S. Quantification of airborne Mycobacterium tuberculosis in health care setting using real-time qPCR coupled to an air-sampling filter method. Aerosol Sci Technol. 2005;39(4):371–376. doi:10.1080/027868290945767.
57. Hubad B, Lapanje A. Inadequate hospital ventilation system increases the risk of nosocomial Mycobacterium tuberculosis. J Hosp Infect. 2012;80(1):88–91. doi:10.1016/j.jhin.2011.10.014
58. Matuka O, Singh TS, Bryce E, et al. Pilot study to detect airborne Mycobacterium tuberculosis exposure in a South African public healthcare facility outpatient clinic. J Hosp Infect. 2015;89(3):192–196. doi:10.1016/j.jhin.2014.11.013
59. Sornboot J, Aekplakorn W, Ramasoota P, Bualert S, Tumwasorn S, Jiamjarasrangsi W. Detection of airborne Mycobacterium tuberculosis complex in high-risk areas of health care facilities in Thailand. Int J Tuberc Lung Dis. 2019;23(4):465–473. doi:10.5588/ijtld.18.0218
60. Li H, Bai R, Zhao Z, et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci Rep. 2018;38(6):BSR20181170. doi:10.1042/BSR20181170
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
