Back to Journals » Clinical Interventions in Aging » Volume 12
Apolipoprotein ε7 allele in memory complaints: insights through protein structure prediction
Authors Youn YC , Lim YK, Han SH , Giau VV , Lee MK, Park KY, Kim SY , Bagyinszky E, An SSA , Kim HR
Received 27 December 2016
Accepted for publication 25 March 2017
Published 11 July 2017 Volume 2017:12 Pages 1095—1102
DOI https://doi.org/10.2147/CIA.S131172
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Walker
Young Chul Youn,1,* Yong Kwan Lim,2,* Su-Hyun Han,1 Vo Van Giau,3 Mi-Kyung Lee,2 Kwang-Yeol Park,1 SangYun Kim,4,5 Eva Bagyinszky,3 Seong Soo A An,3 Hye Ryoun Kim2
1Department of Neurology, 2Department of Laboratory Medicine, Chung-Ang University College of Medicine, Seoul, 3College of BioNano Technology, Gachon BioNano Research Institute, Gachon University, 4Department of Neurology, Seoul National University Bundang Hospital, 5Department of Neurology, Seoul National University College of Medicine, Seongnam, South Korea
*These authors contributed equally to this work
Purpose: APOE ε7 gene is a rare mutant form of APOE ε3. The mutation occurs in the lipid-binding domain of APOE. Based on the protein’s structure, APOE ε7 is expected to function in lipid and β-amyloid metabolism, similar to APOE ε4. However, unlike that for APOE ε4, the mechanisms responsible for Alzheimer’s disease (AD) cases associated with APOE ε7 expression have not been elucidated. The present study aims to investigate the association between APOE ε7 expression and cognitive impairment.
Methods: APOE was sequenced in DNA samples collected from 344 memory-complaint patients who visited the memory clinic, and from 345 non-memory-complaint individuals from the health promotion center. The protein structures of ApoE3, ApoE4, and ApoE7 were predicted.
Results: Three ε3/ε7 heterozygote individuals who were all classified under the memory-complaint group were identified. Of these, two subjects were clinically diagnosed with AD with small vessel disease, and the remaining individual was diagnosed with subjective cognitive impairment. This study predicted the protein structures of ApoE3, ApoE4, and ApoE7 and determined the three-dimensional structure of the carboxy terminus of ApoE7, which participates in an electrostatic domain interaction similar to that of APOE ε4. APOE K244 or K245 mutations for APOE ε7 were not found in the Korean reference genome database, which contains information (http://152.99.75.168/KRGDB/browser/mainBrowser.jsp) from 622 healthy individuals.
Conclusion: As verified by the results of structural prediction, APOE ε7 could serve as another risk factor for cognitive impairment and is particularly associated with vascular disease. However, additional studies are required to validate the pathogenic nature of APOE ε7.
Keywords: apolipoprotein structure, Alzheimer’s disease, vascular cognitive impairment, small vessel disease
Introduction
Alzheimer’s disease (AD) is the most common form of dementia; it is characterized by progressive impairment in memory and other cognitive functions, which can be accompanied by additional symptoms, including behavioral changes. AD can be classified as either early onset (<65 years of age) or late onset (>65 years of age). The APOE ε4 gene allele has been found to act as a primary risk factor for both late- and early-onset AD.1,2 ApoE has been demonstrated to play a key role in lipid metabolism and is also involved in neuronal growth and development and in maintaining synaptic integrity. The APOE gene that encodes the ApoE protein is located in the long arm of chromosome 19. ApoE is a major cholesterol carrier that facilitates lipid transport. Alleles ε2, ε3, and ε4 of ApoE are the most common sites for ApoE single-nucleotide polymorphism variations. ApoE3 contains a cysteine at position 112 and an arginine at position 158, whereas ApoE4 and ApoE2 contain arginine and cysteine residues at positions 112 and 158, respectively.2
APOE ε4 is associated with increased AD risk, but APOE ε2 is associated with reduced AD risk and later disease onset than APOE ε3. APOE ε3 is the most common allele in the human population and can naturally undergo mutations into the APOE ε7 variant, wherein two lysine residues replace glutamic acid residues at positions 244 (rs140808909) and 245 (rs190853081) in the carboxyl terminus, in which the major lipid-binding region is located (residues 244–272).3
The APOE ε7 mutant is associated with hyperlipidemia and atherosclerosis and is reported to bind defectively to the low-density lipoprotein (LDL) receptor, to bind with high affinity to heparin, and like APOE ε4, to associate preferentially with very low-density lipoprotein (VLDL).4 Therefore, it was hypothesized that similar to APOE ε4, APOE ε7 is a risk factor for AD, especially if it occurs with vessel disease. So far, there have been no reports of APOE ε7 in the context of AD; the present study identified a female APOE ε7 heterozygote with an unreported case of AD with small vessel disease. It was hypothesized that the structure of this individual’s ApoE protein could be similar to that of ApoE4, which interferes with lipid5,6 and β-amyloid metabolism,7 thereby contributing to pathogenesis, leading to cognitive impairment.
Several methods have been developed to determine APOE genotypes, such as polymerase chain reaction (PCR)-restriction fragment length polymorphism,8–10 PCR-single strand conformation polymorphism,11,12 and real time PCR.13 Considering that these methods can misidentify APOE ε7 alleles as ε3, an evaluation was performed via sequencing analysis to avoid such errors.13
The present study validated the frequency of APOE ε7 occurrence in subjects who visited the memory clinic because of memory complaints and in members of the general population who visited the health promotion center with no memory complaints and discussed their clinical significance herein.
Materials and methods
To verify the frequency of APOE ε7, genomic DNA was extracted from stored blood samples following standard procedures. All study subjects provided written informed consent allowing genetic and clinical data to be used for research purposes. This study was conducted with approval from the Institutional Review Board of Chung-Ang University Hospital (Approval No C2015255).
Subjects
The subjects of this study included 745 people who visited the memory clinic or the health promotion center at the Chung-Ang University Hospital between June 2003 and December 2004 and had already requested APOE genotyping. Out of 745 total visitors, 400 individuals visited the memory clinic, in which 344 were categorized under the amnesia group, and 56 were excluded from the study because they did not have memory complaints (Figure 1). The amnesia group comprised subjects who complained of memory impairments, including dementia, mild cognitive impairment, and subjective memory impairment. Patients with memory complaints were evaluated by a neurologist, who obtained detailed medical history and performed neurological examinations, routine laboratory tests, and magnetic resonance imaging (MRI). A standardized mental status battery was also conducted, which included Korean Mini-Mental State Examination (MMSE), Clinical Dementia Rating (CDR) and CDR Sum of Boxes, Hachinski ischemic scale, geriatric depression scale, and Seoul Neuropsychological Screening Battery.14
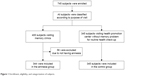  | Figure 1 Enrollment, eligibility, and categorization of subjects. |
The remaining 345 control subjects underwent routine health checkups and did not complain of memory problems.
APOE genotyping and sequencing analysis
Genomic DNA was isolated from peripheral blood samples, using Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA). APOE genotyping was performed using the Multiplex Amplification Refractory Mutation System (ARMS), as previously described.15 Briefly, the following primer sets were used: Cys158/Arg158 (5′-ATGCCGATGACCTGCAGAATT-3′)/(5′-ATGCCGATGACCTGCAGAATC-3′); Cys112/Arg112 (5′-CGCGGACATGGAGGACGTTT-3′)/(5′-CGCGGACATGGAGGACGTTC-3′); and common primer (5′-GTTCAGTGATTGTCGCTGGGCA-3′). The common primer was paired with Arg158/Cys158 or Arg112/Cys112 and had amplicon lengths of 588 and 451 bp, respectively. For the internal positive control, the 360-bp fragment of the α1-antitrypsin gene was co-amplified. The following PCR profile was used for amplification: initial denaturation at 95°C for 4 minutes; 35 cycles of denaturation at 96°C for 45 seconds, annealing at 66°C for 45 seconds, and extension at 72°C for 45 seconds; and final extension at 72°C for 5 minutes. Amplicons were separated via 1% agarose gel electrophoresis at 100 V for 40 minutes, and DNA was stained with ethidium bromide. Multiplex ARMS method was designed to discriminate only among APOE ε2, ε3, and ε4 species; because of sequence homology with the target sequence in multiplex ARMS, APOE ε7 was assumed to be misidentified as APOE ε3. Therefore, an additional PCR step was performed to distinguish between APOE ε7 and APOE ε3. The oligonucleotide primer E9 (5′-CGTGAATTCGCATGGCTGCAGGCTTCGGCGTTC-3′) was used as a common primer. Two specific primers, namely, primer A/A (5′-GTGCGCGCCAAGCTGAAGA-3′) and primer G/G (5′-GTGCGCGCCAAGCTGGAGG-3′), were paired with the common primer E9 to generate 217-bp amplicons.5 The following PCR profile was used for amplification: initial denaturation at 96°C for 10 minutes; 30 cycles at 96°C for 30 seconds, 65°C for 30 seconds, and 72°C for 60 seconds; and final extension at 72°C for 10 minutes. PCR products were then electrophoresed as described above (Figure 2). To verify the correct APOE ε7 allele, the nucleotide sequence of the APOE gene was analyzed using the E7 primer (5′-CGTGAATTCGCATGGCTGCAGGCTTCGGCGTTC-3′) and the common primer E9, which covered exon 4 of the APOE.5,16–18
Statistical analysis and structural prediction of ApoE7
The chi-squared test was performed to compare differences in the distribution of gender, APOE genotype, and allele frequency among the subjects. The Mann–Whitney U-test was performed to compare the median age between the amnesia group and control group. Statistical significance was considered at P-value <0.05. All statistical analyses were performed using SPSS Version 19.0 (IBM Corp, Armonk, NY, USA).
The three-dimensional (3D) structures of normal (ApoE3) and mutant (ApoE7) forms were generated using the RaptorX web server (http://raptorx.uchicago.edu/), a protein structure prediction server that takes amino acid sequences as inputs. Discovery Studio 3.5 Visualizer from Accelrys (San Diego, CA, USA) was used to display the attractive interactions between the relevant regions of the APOE protein (amino acids 1–299).
Results
Demographics in amnesia and control groups
The demographic features of a total of 689 subjects are summarized in Table 1. Whereas male and female subjects were evenly distributed in the control group, female patients were twice as many as the male patients in the amnesia group. The median age for subjects in the amnesia group was 70 years (69 years for males, 71 years for females), which was higher than that of the control group.
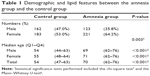  | Table 1 Demographic and lipid features between the amnesia group and the control group |
APOE genotype frequencies
The frequencies of the genotypes and alleles of the APOE genes in the amnesia group and control group are compared in Table 2. In the control group, the genotype frequency and allele frequency were very similar to those previously reported in the literature.19 The amnesia group showed a higher ε3/ε4 frequency and lower ε3/ε3 frequency than those of the control group. The APOE ε4 allele frequency in the amnesia group was twice that of the control group. Among all subjects, only three individuals were identified to carry the APOE ε7 allele, all of which were ε3/ε7 heterozygotes. All three cases belonged to the amnesia group.
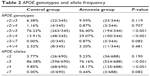  | Table 2 APOE genotypes and allele frequency |
The first case was an 80-year-old female who visited the memory clinic because of forgetfulness regarding a recent event, slowness of thought, and delusions. Her daughter reported that she suffered from loss of energy, attention deficit, poor hygiene, and apathy, which had started 2 years prior and had progressed slowly. Her MMSE score was 11/30, the CDR was 2, and the score for the geriatric depression scale was 18/30. Results of brain MRI reported moderate to severe white matter hyperintensities in both the periventricle and centrum semiovale, as well as multiple lacunar infarctions with diffuse brain atrophy. Clinical diagnostic impression was subcortical ischemic vascular dementia; however, the possibility of AD dementia with small vessel disease cannot be ruled out.
The second case was a 76-year-old man, who was referred to the memory clinic because of memory loss, frequent occurrences of getting lost, and poor verbal expression within the last 12 months. The patient was suffering from hypertension for the past 10 years. His MMSE score was 25/30 and CDR was 1. Results of formal neuropsychological assessment reported naming difficulty, visuospatial dysfunction, and impaired verbal memory. Brain MRI showed hyperintensities in both periventricular and deep white matter and diffuse brain atrophy with hippocampal atrophy. The first diagnostic impression was AD with small vessel disease.
The third case was a 61-year-old female managing a clothing store. She visited the memory clinic because of slowly progressive memory lapses over the past year concerning recent facts, prices, and sizes of clothes. In addition, she complained that she often misplaced things in her store and had difficulties with multitasking and calculations. However, no interference with daily functioning was reported. There was no family history of dementia or memory loss, and neurological evaluation revealed no abnormalities. Her MRI results were normal and showed no hippocampal atrophy. Her MMSE score was 29/30, and a formal neuropsychological assessment showed normal performance. She was diagnosed with having subjective memory complaints. Thus, among the analyzed individuals carrying the APOE ε7 allele, two had AD with small vessel disease, but one case was diagnosed with subjective memory complaints. In contrast, no one in the control group had the APOE ε7 allele.
This study also attempted to determine the APOE ε7 allele frequency in the Korean reference genome database (KRGD) of Center for Disease Control and Prevention (http://152.99.75.168/KRGDB/browser/mainBrowser.jsp), which contains genetic information on 622 healthy individuals and 32 million variants. Neither APOE K244 nor K245 was found in this database.
Comparison between the predicted structures of ApoE3 and ApoE7
Figure 3 shows the protein structures of ApoE3, ApoE7, and ApoE4 generated by the RaptorX 3D prediction program. Both ApoE3 and ApoE7 demonstrate an attractive electrostatic interaction between the residues Arg61 and Glu255. Whereas ApoE3 shows a repulsion between Glu49-50 and Glu244-245, ApoE7 has an attractive interaction between Glu49-50 and Lys244-245, which is due to the Glu → Lys mutations at positions 244 and 245 located in the major lipid-binding region of ApoE (residues 244–272). Similar to ApoE3, ApoE4 has an attractive interaction between Arg61 and Glu255 and a repulsive interaction between Glu49-50 and Glu244-245; however, in ApoE4, Arg112 participates in an attractive interaction with Glu255.
  | Figure 3 Ribbon diagram depicting ApoE isoform-specific differences in domain interactions. |
Discussion
The ApoE protein plays a crucial role in cognitive function, and the APOE genotype is one of the risk factors for AD. The association between AD and APOE ε4 has been well established,20 but alleles like APOE ε7 remain to be fully investigated. In this study, the APOE ε7 allele was identified only in patients under the memory-complaint group; other patients in the medical center were categorized under the control group. In addition to 345 control subjects, a search was also carried out for the occurrence of the APOE ε7 allele in the KRGD (http://152.99.75.168/KRGDB/browser/mainBrowser.jsp); however, no variants at positions 244 and 245 were found to correspond to APOE ε7. Consequently, no APOE ε7 allele was identified in the 967 control subjects. Two individuals in the memory-complaint group harboring the APOE ε7 showed features that were clinically compatible with AD with small vessel disease. Given that the third case was diagnosed as subjective cognitive impairment and the individual was relatively young with normal cognition based on the formal neuropsychological test, a periodic follow-up on the case has been planned. Despite the small sample size, analysis of the cases suggests the possibility that APOE ε7 polymorphism is associated with cognitive impairment.
ApoE plays critical roles in various biological processes, including transport of cholesterol and cholesterol-like molecules, such as β-amyloid. ApoE has two defined structural domains, namely, an N-terminal domain (residues 1–191) containing the LDL receptor-binding site (residues 136–150) and a C-terminal domain (residues 216–299) containing the major lipid-binding site (residues 244–272) (Figure 3).21 The ApoE protein is encoded by the APOE gene, which is located on chromosome 19. ApoE has the following three well-studied alleles: APOE ε2 (frequency in populations, 5%–10%), APOE ε3 (60%–70%), and APOE ε4 (15%–20%).21 These study results also showed the highest APOE ε3 and lowest APOE ε2 frequencies in this study population.
The ApoE isoforms differ only at residues 112 and 158. ApoE3 has Cys112 and Arg158; on the other hand, ApoE4 has arginine in both sites, whereas ApoE2 has cysteines in those sites. In the ApoE4 variant, Arg112 allows the side chain of Arg61 to extend away from the helical bundle,22 resulting in an orientation different from the side chain of Arg61 in ApoE3 and ApoE2.23 Consequently, ApoE4 domain interaction may be influenced by electrostatic binding between Arg61 and Glu255 and between Arg112 and Glu255 (Figure 3).
ApoE7 is a naturally occurring mutant of ApoE3 in humans, with two lysine residues replacing glutamic acid at positions 244 and 245 in the carboxyl terminus. Some studies have suggested that the ApoE7 variant is associated with hyperlipidemia and atherosclerosis; furthermore, ApoE7 is reported to bind defectively to the LDL receptor and to associate preferentially with VLDL.4 Although ApoE7 is a variant of ApoE3, analysis using the RaptorX 3D prediction program showed that the tertiary structure of the ApoE7 carboxy terminus is more similar to that of ApoE4 than ApoE3; as represented in E4 and E7 of Figure 3, these changes affect the conformation and orientation of their two primary domains. The electrostatic interaction between the amino-terminal and carboxy-terminal domains of ApoE7 may be stronger than that of ApoE3 and more similar to that of ApoE4. Furthermore, the two glutamic acid residues at positions 244 and 245 of the C terminus of ApoE7 correspond to residues 244–272 of the carboxy terminal of ApoE3, which is in the major lipid-binding region of ApoE. This change may directly influence the lipid-binding properties of ApoE7.
One study suggested that the contribution of APOE ε4 to the risk of vascular cognitive impairment was independent of other vascular risk factors, including hypertension, dyslipidemia, and atherogenesis.24 A recent meta-analysis has shown that APOE ε4 increases the risk of vascular dementia than APOE ε3;25 therefore, the small vessel disease diagnosed in subjects complaining of memory impairment could be partially explained by the presence of the APOE ε7 mutation.
β-amyloid is the protein primarily linked to AD and has been investigated because of its interaction with ApoE. Many in vitro studies have demonstrated that ApoE binds to the oligomeric form of β-amyloid; however, the ApoE-binding site and the binding mechanisms remain unclear. ApoE3 and ApoE4 bind to β-amyloid at a different rate and influence β-amyloid aggregation.26–28 Analysis of the lipid-binding region at positions 244–272 indicates that ApoE4, preferentially in its oligomeric form,29 is involved in β-amyloid binding.30 ApoE7 is also similar to ApoE4 in terms of electrostatic interaction properties and tertiary protein structure, which could be expected to contribute to the AD pathomechanism; however, this topic requires further investigation.
Limitations
The current study had certain limitations. First, AD diagnoses in the three cases were not confirmed using compatible biomarkers, such as amyloid positron emission tomography or cerebrospinal fluid β-amyloid. Diagnosis relied only on the clinical course and brain MRI results, in which two cases were diagnosed with AD and one was diagnosed with subjective cognitive impairment. Given that the analysis was performed using stored DNA samples and retrospective review of the patients’ clinical records, an additional biomarker study was not performed on these patients. The present study screened APOE ε7 in 689 (344 memory complaints and 345 control) subjects and additionally searched the variant in position 622 in the KRGD. Consequently, results indicate that APOE ε7 is present only in the memory-complaint group and not in control subjects and the KRGD database. Considering that APOE ε7 is not a common variant, further studies should be conducted with a larger sample size with well-characterized AD patients. In addition, future studies should validate the pathogenic properties of APOE ε7.
Conclusion
In the present study, subjects visiting the medical center with the APOE ε7 variant were identified only in the memory-complaint group. The predicted protein structure of ApoE7 in these individuals shows an electrostatic domain interaction that is similar to that of ApoE4. These findings suggest the possibility that APOE ε7 is another risk factor for cognitive impairment, especially in cognitive diseases that are associated with vascular disease. Therefore, the APOE ε7 allele must also be considered in performing APOE genotyping in dementia evaluation. However, additional studies are required to validate the pathogenic role of APOE ε7 in cognitive impairment, particularly in AD.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education (NRF-2017R1A2B4011631) and a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3331).
Ju-Won Seok (Department of Nuclear Medicine, Chung-Ang University College of Medicine) performed the neuroimaging. Kyu Hwan Shim (College of BioNano Technology, Gachon BioNano Research Institute) performed tests and analyzed the ApoE7 data.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Tang MX, Maestre G, Tsai WY, et al. Relative risk of Alzheimer disease and age-at-onset distributions, based on APOE genotypes among elderly African Americans, Caucasians, and Hispanics in New York City. Am J Hum Genet. 1996;58(3):574–584. | ||
Laws SM, Hone E, Gandy S, Martins RN. Expanding the association between the APOE gene and the risk of Alzheimer’s disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J Neurochem. 2003;84(6):1215–1236. | ||
Dong J, Balestra ME, Newhouse YM, Weisgraber KH. Human apolipoprotein E7:lysine mutations in the carboxy-terminal domain are directly responsible for preferential binding to very low density lipoproteins. J Lipid Res. 2000;41(11):1783–1789. | ||
Yamamura T, Dong LM, Yamamoto A. Characterization of apolipoprotein E7 (Glu244→Lys, Glu245→Lys), a mutant apolipoprotein E associated with hyperlipidemia and atherosclerosis. J Lipid Res. 1999;40(2):253–259. | ||
Yanagi K, Yamashita S, Hiraoka H, et al. Increased serum remnant lipoproteins in patients with apolipoprotein E7 (apo E Suita). Atherosclerosis. 1997;131(1):49–58. | ||
Li H, Dhanasekaran P, Alexander ET, Rader DJ, Phillips MC, Lund-Katz S. Molecular mechanisms responsible for the differential effects of ApoE3 and ApoE4 on plasma lipoprotein-cholesterol levels. Arterioscler Thromb Vasc Biol. 2013;33(4):687–693. | ||
Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240(4852):622–630. | ||
Senanarong V, Harnphadungkit K, Lertrit P, et al. Experience of ApoE study in Thai elderly. J Med Assoc Thai. 2001;84(2):182–187. | ||
Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31(3):545–548. | ||
Kontula K, Aalto-Setala K, Kuusi T, Hamalainen L, Syvanen AC. Apolipoprotein E polymorphism determined by restriction enzyme analysis of DNA amplified by polymerase chain reaction: convenient alternative to phenotyping by isoelectric focusing. Clin Chem. 1990;36(12):2087–2092. | ||
Tsai MY, Suess P, Schwichtenberg K, et al. Determination of apolipoprotein E genotypes by single-strand conformational polymorphism. Clin Chem. 1993;39(10):2121–2124. | ||
Wilton S, Lim L. Rapid identification of ApoE alleles by multiple-single-strand conformation polymorphism (SSCP) analysis. Trends Genet. 1995;11(9):341. | ||
Park C, Lee S, Ki C, Kim J. Discrepancy in genotyping of apolipoprotein E between allele-specific PCR and fluorescence resonance energy transfer or sequencing. Korean J Lab Med. 2010;30:325–328. | ||
Kang Y, Na DL. Seoul Neuropsychological Screening Battery (SNSB). 1st ed. Incheon: Human Brain Research & Consulting Co; 2003. | ||
Donohoe GG, Salomaki A, Lehtimaki T, Pulkki K, Kairisto V. Rapid identification of apolipoprotein E genotypes by multiplex amplification refractory mutation system PCR and capillary gel electrophoresis. Clin Chem. 1999;45(1):143–146. | ||
Emi M, Wu LL, Robertson MA, et al. Genotyping and sequence analysis of apolipoprotein E isoforms. Genomics. 1988;3(4):373–379. | ||
Matsunaga A, Sasaki J, Komatsu T, et al. A novel apolipoprotein E mutation, E2 (Arg25Cys), in lipoprotein glomerulopathy. Kidney Int. 1999;56(2):421–427. | ||
Hagiwara M, Yamagata K, Matsunaga T, et al. A novel apolipoprotein E mutation, ApoE Tsukuba (Arg 114 Cys), in lipoprotein glomerulopathy. Nephrol Dial Transplant. 2008;23(1):381–384. | ||
Kim KW, Jhoo JH, Lee KU, et al. Association between apolipoprotein E polymorphism and Alzheimer’s disease in Koreans. Neurosci Lett. 1999;277(3):145–148. | ||
Weisgraber KH, Mahley RW. Human apolipoprotein E: the Alzheimer’s disease connection. FASEB J. 1996;10(13):1485–1494. | ||
Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc Natl Acad Sci U S A. 2006;103(15):5644–5651. | ||
Dong LM, Wilson C, Wardell MR, et al. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J Biol Chem. 1994;269(35):22358–22365. | ||
Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252(5014):1817–1822. | ||
Prince M, Lovestone S, Cervilla J, et al. The association between APOE and dementia does not seem to be mediated by vascular factors. Neurology. 2000;54(2):397–402. | ||
Yin YW, Li JC, Wang JZ, et al. Association between apolipoprotein E gene polymorphism and the risk of vascular dementia: a meta-analysis. Neurosci Lett. 2012;514(1):6–11. | ||
Strittmatter WJ, Weisgraber KH, Huang DY, et al. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(17):8098–8102. | ||
Webster S, Rogers J. Relative efficacies of amyloid beta peptide (A beta) binding proteins in A beta aggregation. J Neurosci Res. 1996;46(1):58–66. | ||
Wood SJ, Chan W, Wetzel R. An ApoE-Abeta inhibition complex in Abeta fibril extension. Chem Biol. 1996;3(11):949–956. | ||
Carter DB. The interaction of amyloid-beta with ApoE. Subcell Biochem. 2005;38:255–272. | ||
Liu Q, Wu WH, Fang CL, et al. Mapping ApoE/Aβ binding regions to guide inhibitor discovery. Mol Biosyst. 2011;7(5):1693–1700. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

