Back to Journals » International Journal of General Medicine » Volume 13
Antimicrobial Susceptibility Testing and Phenotypic Detection of MRSA Isolated from Diabetic Foot Infection
Authors Anwar K , Hussein D , Salih J
Received 17 September 2020
Accepted for publication 5 November 2020
Published 2 December 2020 Volume 2020:13 Pages 1349—1357
DOI https://doi.org/10.2147/IJGM.S278574
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Khanda Anwar,1 Dlsoz Hussein,2 Jamal Salih3,4
1Microbiology Department, College of Medicine, University of Sulaimani, Sulaymaniyah, Kurdistan Region, Iraq; 2Microbiology Department, Central Public Health Laboratory 1, Sulaymaniyah, Kurdistan Region, Iraq; 3Physiology Department, College of Medicine, University of Sulaimani, Sulaymaniyah, Kurdistan Region, Iraq; 4Diabetes and Endocrine Centre, Sulaymaniyah, Iraq
Correspondence: Jamal Salih
Physiology Department, College of Medicine, University of Sulaimani, New–Street-27, Zone 209, P.O. Box: 334, Sulaimaniyah, Kurdistan Region, Iraq
Tel +00964-7702441985
Email [email protected]
Background: Diabetic foot infection (DFI) is a common and costly complication of diabetes that may be caused by various bacteria with multi-resistant genes. The aim of this study is to evaluate the efficacy of phenotypic methods for identification of methicillin-resistant Staphylococcus aureus (MRSA) with genotypic detection of MRSA-related genes.
Methods: In this cross-sectional study, swab samples were collected from patients with DFI from hospitals in Sulaimani/Iraq in April–July 2019. All the samples were processed for microbiological assessment and further MRSA phenotypic and genotypic testing.
Results: A total of 46 swab samples were collected from diabetic foot ulcers of 29 males and 17 females. Most samples (93.5%) showed positive growth, with higher proportions of monomicrobial (23; 53.5%) than mixed-bacterial infections (20; 46.5%) and S. aureus as the predominant pathogen. Conventional methods of MRSA detection, such as cefoxitin disc diffusion, can predict methicillin resistance in 45.8% of the cases. Real-time/conventional PCR showed that 41.6% of Staphylococcus aureus were positive for the mecA gene, while none of the isolates was positive for PVL.
Conclusion: Staphylococcus aureus was the predominant pathogen in DFI. Although cefoxitin and oxacillin disc diffusion methods can help in the prediction of MRSA, real-time PCR is a reliable method for MRSA detection and confirmation.
Keywords: diabetic foot, infection, MRSA, genotypic detection
Introduction
Diabetes mellitus is a common chronic disease, characterized by persistent hyperglycemia. One of the most serious complications of this disease is diabetic foot infection (DFI),1 which is caused by single or multiple microorganisms.2 Aerobic Gram-positive cocci, such as Staphylococcus aureus, are the predominant organism responsible for acute DFI. However, polymicrobial isolates are mostly observed in chronic wound infections. Numerous studies have reported that DFI was caused by multidrug-resistant organisms, such as extended-spectrum β-lactamase producing Gram-negative rods and Methicillin-resistant Staphylococcus aureus (MRSA).3,4
The presence of MRSA strains is focused on the presence of mecA, which determines the synthesis of abnormal penicillin-binding proteins (PBPs). These PBPs normally have a strong affinity for the β-lactam ring5; however, in the strains of MRSA, another PBP2a has a very low affinity for binding to the β-lactam antibiotics. This leads to the methicillin antibiotics failing to destroy the bacterial cell wall.6
MRSA detection commonly depends on a combination of disc diffusion and molecular methods. The phenotypic (cefoxitin and oxacillin disc diffusion) method is commonly used, whereas the genotypic method for detecting mecA is highly conserved to MRSA. Additionally, Panton–Valentine leukocidin (PVL), which is a cytolytic toxin produced by less than 5% of S. aureus strains, is regarded as a virulence factor; however, its role in DFI remains controversial.7 Since the phenotypic method is widely available, it remains the method of choice, but phenotypic method is time-consuming, and the strain detection can be affected by environmental factors. Therefore, detection of the mecA gene and penicillin-binding proteins by molecular techniques is considered for MRSA confirmation.8
Treatment of MRSA infections is a global challenge. Nowadays, MRSA provides resistance to the previously effective β-lactam antibiotics.9 Glycopeptides inhibit bacterial cell wall synthesis using a different mechanism to that of the β-lactams, so they are potentially active against all S. aureus strains, including MRSA.10 Vancomycin and teicoplanin are highly effective in treatment of serious MRSA infections; however, the suboptimal response often necessitates the addition of a second or third antimicrobial agent, such as rifampicin and aminoglycosides.11,12 Reasonable empirical antibiotic treatments in limb-threatening infections include a combination of an anti-anaerobic agent like clindamycin and an anti-aerobic antibiotic.13 Over the past few years, new cephalosporins with improved activity against Staphylococcus aureus have been discovered; however, their antimicrobial activities are insufficient to eradicate MRSA infection,14 except for the fourth generation parenteral cephalosporin, which has shown good antibacterial activity against MRSA.
In our locality, data on DFI are lacking, and there is neither a sufficiency of published studies nor consistent practical, evidence-based guidance to suggest appropriate antibiotic choices based on local data. The aim of this study is to evaluate the efficacy of phenotypic methods for identification of MRSA. An additional aim was genotypic detection of MRSA-related genes (mecA and PVL).
Methodology
This study was conducted in different hospitals in Sulaimani and ethical approval was obtained from the Directorate of Health in Sulaimani. This cross-sectional study included 46 patients with type II diabetes mellitus presented with DFI in Diabet Center, Shar Hospital and Surgical Teaching Hospital in Sulaimani from April to July 2019. This study was conducted in accordance with the Declaration of Helsinki and signed informed consent was obtained from all patients before enrollment. The swab samples were selected according to the presence of signs and symptoms of foot infection.13 The subjects were classified into two groups (“hospitalized” and “community”), based on admission time to the hospital: patients who had been seen in outpatient clinics and admitted to hospitals within 48 hours were classified as non-hospitalized (community group) patients, while those who stayed in hospital for more than 48 hours were recorded as hospitalized patients.15 From each patient, random blood sugar, HbA1c, and demographic data such as age, gender, smoking, and alcoholic status were obtained.
All DFI wounds were selected to be tested by taking swabs.16 The swabs were promptly delivered to the microbiology laboratory in Emergency and Plastic Surgery Hospital in Sulaimani within 2 hours from collection or stored at 4°C for 4 hours; subsequently, the samples were processed17 by routine standard culture techniques and prompt identification and freezing of the samples for molecular work.18 Antibiotic susceptibility testing was done by the Kirby–Bauer disc diffusion method, in accordance with the guidelines of the National Committee for the Clinical Laboratory Standards Institute (CLSI).19
Phenotypic Detection of MRSA
All isolated S. aureus strains were tested by oxacillin and cefoxitin disc diffusion methods, according to what was described previously.19
Genotypic Detection of MRSA
Two genes (mecA and PVL) were selected in this study to be analyzed by molecular techniques. Bacterial reactivation18 and DNA extraction were performed on all isolated coagulase-positive S. aureus strains (MRSA and MSSA) according to the recommendation of the manufacturers of the Automated DNA extraction kit (MagCore® Nucleic Acid Extraction Kit RBC bioscience-Taiwan).
Two sets of primers were used in this study, designed by Macrogen/Korea, both forward and reverse (Table 1), according to what was previously tested by Bhatta et al (2016). Main stock primers (100 pmol/µL) were prepared by suspending each primer (forward and reverse) in free DNA and RNA injection water (for mecA1 and mecA2 oligonucleotide by adding 300 µL, for LUK-PV-1270 µL, and for LUK-PV-2290 µL), according to what was performed.20 Table 2 illustrates the base pair sequences of both primers.
 |
Table 1 Primer (mecA, PVL) Gene Oligonucleotide Sequence |
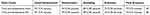 |
Table 2 Real-Time PCR Conditions |
Polymerase Chain Reaction (Real-Time PCR)
The presence of mecA and PVL was determined among all isolated strains of S. aureus; mecA and PVL genes were analyzed by real-time PCR according to a specific condition (Table 2) that was programmed in the protocol by Bhatta et al, and PCR amplification and real-time hybridization were conducted using the Promega GoTaq® Probe qPCR Master Mix Kit (USA). Mastermix contains GoTaq® Hot Start Polymerase, SYBR green dye, MgCl2, dNTPs, and a proprietary reaction buffer. DNA samples to be run in PCR were diluted to10 ng/µL.20
Multiplex conventional PCR was used for detection of mecA and PVL for all isolated S. aureus strains (24) by using the same primer as in Table 1, but with different PCR conditions, programmed in the protocol by Bhatta et al (2016) and illustrated in Table 3.
 |
Table 3 Multiplex PCR Conditions for Amplification of mecA and PVL |
Gel Electrophoresis
Agarose gel electrophoresis was prepared according to what was prepared previously.21 All PCR products were run on 1% agarose gel electrophoresis for 30 minutes and 100 bp DNA ladder or marker was used for the validation of the length of the amplified PCR product. The gel image was visualized using UV light and photographed.
Statistical Analysis
All the data were analyzed using Microsoft Excel for statistical analysis. Mean, standard deviation, and chi-square tests were used to correlate different parameters, and Fischer exact tests were used to find significant values; p < 0.05 was used as a level of significance in this study.
Results
In this study, 46 swab samples from DFI were investigated. The participants were 29 males (63%) and 17 females (37%), with a male-to-female ratio of 1.7:1. The participant’s age ranged from 41 to 82 years, with a mean of 57.4 (±SD8.6). A majority of the samples were collected from outpatient clinics (33; 71.7%), and the rest (13; 28.3%) were from hospitalized patients.
Glucose control status was assessed; the mean HbA1c was 8.4% (±SD 1.9), while the mean random blood sugar was 236.2 mg/dl (±SD 102.3). According to the American Diabetes Association’s general glycemic target, 34 patients (74%) had HbA1c above the target (7%), while 12 (26%) had last HbA1c of ≤7%.1
Lower limb or foot amputation was observed in six participants (13%), and 14 patients (30.5%) had foot deformities. Smoking status, alcohol consumption, and previous antibiotic intake were illustrated in Table 4, and it is obvious that current smoking and alcohol consumption was less prevalent among participants.
 |
Table 4 Characteristics of the Study Participants |
Out of 46 swab samples, 43 (93.5%) showed positive bacterial growth, while only three samples (6.5%) were found to be negative for bacterial growth. The relation between HbA1c and positive culture result was analyzed and found to be statistically insignificant.
Of 43 positive cultures, 31 samples (72%) were from outpatient clinics and 12 (28%) were from hospitalized patients. Of positive culture results, 23 patients (53.5%) had monomicrobial growth and 20 (46.5%) had more than one organism isolated from their lesions. Table 5 shows that most of the hospital isolates were monomicrobial and most of the community-based results were polymicrobial, but the difference was statistically insignificant.
 |
Table 5 Distribution of Positive Growth Culture Among Community-Acquired Patients and Hospitalized Patients |
A total of 62 bacteria were isolated according to colonial morphology from 43 growth-positive diabetic patients, resulting in an average of 1.4 organisms per sample. The results showed that Gram-positive organisms (36; 58%) were more prevalent than Gram-negative organisms (26; 42%).
The most common isolated species among Gram-positive bacteria was Staphylococcus aureus (24; 38.7%), based on colonial morphology on blood agar and mannitol salt agar. Among all 24 isolated Staphylococcus aureus strains, mixed infection was observed in nine (37.5%), while 15 (62.5%) were monomicrobial. The majority of S. aureus isolates were from outpatients (17; 70.8%), while seven (29.2%) were reported from hospitalized patients (p value: 0.0223). Table 6 illustrates the relation of this distribution to the culture result.
 |
Table 6 Distribution of S. aureus Among Hospitalized Patients and Community Patients |
Antimicrobial susceptibility pattern was tested for all the 24 isolated S. aureus strains using the Kirby–Bauer disc diffusion method. The sensitivity was interpreted according to CLSI.19 The results were tested against 14 antimicrobial agents and expressed as a percentage of resistance or sensitivity to antimicrobial agents. Penicillin showed the highest resistance (100%) among all the antimicrobial agents, followed by trimethoprim (66.6%), azithromycin (58.3%), and ciprofloxacin and tetracycline (54.2% for each). The sensitivity was highest (100%) to vancomycin, followed by rifampicin (87.5%), gentamicin (83.3%), and chloramphenicol (70.8%). Table 7 illustrates all resistance and sensitivity patterns for all antimicrobial tests.
 |
Table 7 Antimicrobial Susceptibility Pattern of Clinical Staphylococcus aureus Isolated from Diabetic Foot Infections |
Phenotypic Detection of MRSA
Two tests were performed for MRSA detection: cefoxitin and oxacillin disc tests. The results showed that among all isolated S. aureus strains, 11 (45.8%) were cefoxitin-resistant, while nine (37.5%) were oxacillin-resistant, which was statistically significant (p value: 0.00). Table 8 demonstrates the distribution of cefoxitin and oxacillin disc diffusion tests among all isolated S. aureus strains.
 |
Table 8 Cefoxitin and Oxacillin Disc Diffusion Tests |
Genotypic Detection for mecA Gene and PVL Gene
The MRSA gene (mecA) and virulence gene (PVL) were tested by q-PCR, and the results were read by the microsoftware mic PCR program.
mecA Gene Detection
mecA was analyzed by q-PCR for all S. aureus isolates (24). Among all isolated strains, 10 (41.6%) were positive for mecA; Figure 1 illustrates positive results in the Amplification plot (green cycle). A majority of the mecA-positive isolates were obtained from community patients (60%), while the remaining mecA-positive strains (40%) were from hospitalized patients.
 |
Figure 1 Amplification plot of mecA gene (green cycle). |
The sources of samples (hospitalized and community) among diabetic patients in relation to three methods used in diagnoses of MRSA were analyzed, and there was no relation between the test type and the source of the samples (Table 9).
 |
Table 9 MRSA Among Hospitalized and Community Patient Comparison of Phenotypic and Genotypic Methods for the Detection of MRSA |
PVL Gene Detection
PVL was analyzed by q-PCR for all isolated strains of S. aureus (24), and none of the strains was found to be positive for this gene.
Multiplex PCR for mecA and PVL Gene
Both genes (mecA and PVL) were run in a multiplex PCR for all 24 isolated S. aureus strains. An amplification of the mecA gene showed a 310 bp fragment in 10 (41.6%) samples; no positive results for PVL were observed (Figure 2).
Discussion
DFI contributes significantly to morbidity and mortality, which can arise from uncontrolled blood sugar and poor self-care.22 Proper glycemic control is important for infection eradication and ulcer healing.23 Although our participants’ mean glycemia, expressed as HbA1c, was not very high (8.4%), a majority of participants with DFI had HbA1c above the general target level (>7%). In line with previously conducted studies, the mean age of participants was 57.4, and most of the patients were men.24,25 The participants’ lifestyles, professional activities, and jobs might lead to males being more prone to ulcer infection than females.
The microbiological profile of DFI is variable, and it depends on the acute or chronic character of the wound, duration of hospitalization, and previous antibiotic therapy. In this study, most of the diabetic foot ulcers were infected. Although history of antibiotic use was reported for all patients, some patients might not have remembered what medication they took, as there is no medical record in our locality. Similar to Akhi et al (2017),26 the mean number of isolates in this study was 1.4 aerobic bacteria per sample. Our results revealed that more than half of specimens yielded a single isolate, which is in line with Viquez-Molina et al (2018)27 but differs from studies that observed a greater proportion of polymicrobial infection.28,29 The polymicrobial etiology of DFI may be due to depressed immunity in individuals, and poor vascular supply to the feet and chronicity of the wound, which may be contaminated by community-type bacteria and microbial flora. The high prevalence of monomicrobial infections and relatively low rate of isolated pathogens per lesion in this study may be attributable to less severe wound infections.
In agreement with numerous studies, Staphylococcus aureus is the main causative pathogen in diabetic foot ulcer, as we observed S. aureus in 38.7% of the participants. Many studies have exhibited a high prevalence of Gram-positive bacteria in DFI.25,30 Indeed, S. aureus is normal flora of the skin and is the most important true pathogen of skin infections in general. By contrast, other studies have recovered Gram-negative bacteria, Enterobacteriaceae such as E. coli, Proteus spp., and anaerobic bacteria.28,31
The antimicrobial susceptibility test is a useful guide for prescribing appropriate antibiotics for DFI. Resistance to antibiotics is associated with an increased period of hospitalization, high mortality, and increased treatment costs, including a need for alternative medications.32 The initial management of DFI comprises empirical antimicrobial treatment based on the susceptibility data.13 In this study, antibiotic susceptibility was investigated for all the isolated S. aureus using the Kirby–Bauer disc diffusion method; all S. aureus strains were susceptible to vancomycin, which is in agreement with previous studies.26,29,31–33 Undeniably, vancomycin-resistant S. aureus strain was rarely reported.34 Thus, the antibiotic vancomycin was found to be highly effective against Gram-positive organisms, and it still remains the drug of choice for serious infections.
In the current study, the susceptibility of S. aureus to rifampicin, gentamicin, and chloramphenicol was 87.5%, 83.3%, and 70.8%, respectively, meaning that these antibiotics are effective in vitro against S. aureus isolates, as observed by previous studies.26,31,32 Consistently with other studies, we noticed that more than half of the isolated S. aureus were found to be resistant to tetracycline.31 We also noticed that all isolated S. aureus were resistant to penicillin; however, penicillin-sensitive S. aureus was infrequently reported.34 Our results further justified the inappropriateness of penicillin as a treatment option for Staphylococcus aureus infection. It has been revealed that more than 40% of the isolated Staphylococcus aureus from DFI were MRSA; therefore, rapid and accurate detection of MRSA is of major importance for the appropriate clinical management of DFI, including both hospital- and community-based types.26
In the last decade, there have been many attempts by international guidelines such as CLSI to improve and standardize specific phenotypic methods (cefoxitin and oxacillin disc diffusion) for the detection of MRSA.35 However, these two tests are less popularly used by medical laboratories in this region due to less experienced medical staff. In the current study, 45.8% of the isolated S. aureus strains were cefoxitin-resistant, while 37.5% were resistant to oxacillin disc. All the oxacillin-resistant strains were resistant to cefoxitin. These observations were comparable to studies that recorded the same effectiveness of the cefoxitin disc test as a standard phenotypic method.34
In spite of phenotypic diagnosis, nowadays detection of mecA also plays an important role in confirming MRSA. Detection of mecA by real-time PCR to determine MRSA has been considered the standard method because of its accuracy and reproducibility.36 In this study, 41.6% of the isolated S. aureus strains were positive for mecA. Although a single isolate was phenotypically resistant to cefoxitin, it did not show amplification of mecA that might carry another gene, such as mecC, instead of mecA.37 In contrast to this result, mecA was reported among cefoxitin-sensitive strains.33 This variation between the phenotypic and genotypic methods may be related to culture settings, temperature, configuration of culture medium, size of inocula, time of incubation, and manual skill of medical staff.38
According to the oxacillin disc test, two oxacillin-susceptible strains carried mecA and one oxacillin-resistant strain was negative for mecA. This finding may suggest that it is better to use the cefoxitin disc rather than the oxacillin disc test. It has been suggested that the efficacy of the cefoxitin test is superior to that of the oxacillin test and that it sometimes can be used as an alternative to PCR.39 The phenotypic methods of MRSA identification are time-consuming and have their limitations in terms of generating false-positive and false-negative results leading to delay or ineffective antibiotic prescriptions; however, they are still commonly used because of the unavailability and high cost of PCR materials and few experienced staff to carry out PCR techniques.
One of the factors that plays a role in the pathogenicity of Staphylococcus aureus and leads to excess inflammatory responses, tissue damage, and eventually overcoming the host immune response is the presence of PVL.40 However, PVL is less frequently prevalent globally with significant variation in its prevalence among geographical areas (5% in France; 4.9% in the UK).7 In this study, the analysis of PVL was negative, even using conventional and real-time PCR procedure. PVL-negative results were consistently recorded in Poland and Portugal.33,41 It has also been suggested that PVL-positive strains are less frequently detected among DFI.7 Another reason may be the small sample size in our study, and some specimens were obtained from hospital sources, as this gene is mostly prevalent in community species.
We reached the conclusion that accurate detection of the causative agents of DFI is the key step in prescribing effective antibiotics so as to encourage wound healing and minimize subsequent complications. Staphylococcus aureus is the predominant pathogen in DFI. Cefoxitin disc diffusion appears to be a widely available and accurate phenotypic method for MRSA detection and can be helpful in addressing the popularity of resistance pathogens. Indeed, genotypic methods can accurately identify MRSA and its potentially responsible virulent genes; however, PCR is costly, and its use requires special skills. We could not find significant differences between conventional PCR and real-time PCR for mecA detection.
Further studies are recommended and should include samples from different levels of skin lesions in large numbers of patients and analyze virulent factors. It is suggested that the SCCmecA gene be specified, both in community- and hospital-acquired MRSA strains. Analysis of further genes, such as mecC, is needed for strains with positive phenotypic MRSA.
Abbreviations
BMI, body mass index; CLSI, Clinical Laboratory Standards Institute; DFI, diabetic foot infection; FPG, fasting plasma glucose; HbA1c, hemoglobin A1c; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus; PBPs, penicillin-binding proteins; PCR, polymerase chain reaction; PVL, Panton–Valentine leukocidin.
Data and Resource Availability
The datasets generated and analyzed during the current study are available from the corresponding author (J.M, email: [email protected]) upon reasonable request.
Acknowledgment
We would like to thank all the mentioned hospitals and centers from which we collected our study samples.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. There is no prior presentation of this research work.
Disclosure
The authors declare there are no conflicts of interest to disclose.
References
1. Inzucchi SE, Bergenstal RM, Buse JB, et al. Management of hyperglycemia in type 2 diabetes, 2015: a patient-centered approach: update to a position statement of the American Diabetes Association and the European Association for the Study of Diabetes. Diab Care. 2015;38(1):140–149. doi:10.2337/dc14-2441
2. Hinojosa CA, Boyer‐Duck E, Anaya‐Ayala JE, et al. Impact of the bacteriology of diabetic foot ulcers in limb loss. Wound Repair Regeneration. 2016;24(5):923–927.
3. Uckay I, Aragon-Sanchez J, Lew D, Lipsky BA. Diabetic foot infections: what have we learned in the last 30 years? Int J Infectious Diseases. 2015;40:81–91. doi:10.1016/j.ijid.2015.09.023
4. Karmaker M, Sanyal SK, Sultana M, Hossain M. Association of bacteria in diabetic and non-diabetic foot infection–An investigation in patients from Bangladesh. J Infect Public Health. 2016;9(3):267–277. doi:10.1016/j.jiph.2015.10.011
5. Nicoleta M, Gheorghe I, Popa M, et al. Phenotypic and genotypic investigation of resistance and virulence features of methicillin resistant Staphylococcus aureus strains isolated from hospitalized patients. Romanian Biotechnological Letters. 2016;21(3):11591.
6. Malachowa N, DeLeo FR. Mobile genetic elements of Staphylococcus aureus. Cellular Molecular Life Sci. 2010;67(18):3057–3071.
7. Dunyach-Remy C, Ngba Essebe C, Sotto A, Lavigne J-P. Staphylococcus aureus toxins and diabetic foot ulcers: role in pathogenesis and interest in diagnosis. Toxins. 2016;8(7):209. doi:10.3390/toxins8070209
8. Bhutia KO, Singh TS, Biswas S, Adhikari L. Evaluation of phenotypic with genotypic methods for species identification and detection of methicillin resistant in Staphylococcus aureus. Int J Appl Basic Med Res. 2012;2(2):84. doi:10.4103/2229-516X.106348
9. Boudreau MA, Fishovitz J, Llarrull LI, Xiao Q, Mobashery S. Phosphorylation of BlaR1 in manifestation of antibiotic resistance in methicillin-resistant Staphylococcus aureus and its abrogation by small molecules. ACS Infectious Diseases. 2015;1(10):454–459. doi:10.1021/acsinfecdis.5b00086
10. Wang F, Zhou H, Olademehin OP, Kim SJ, Tao P. Insights into key interactions between vancomycin and bacterial cell wall structures. ACS Omega. 2018;3(1):37–45. doi:10.1021/acsomega.7b01483
11. Gould IM, David MZ, Esposito S, et al. New insights into meticillin-resistant Staphylococcus aureus (MRSA) pathogenesis, treatment and resistance. Int J Antimicrob Agents. 2012;39(2):96–104. doi:10.1016/j.ijantimicag.2011.09.028
12. Telles JP, Cieslinski J, Tuon FF. Daptomycin to bone and joint infections and prosthesis joint infections: a systematic review. Brazilian J Infectious Diseases. 2019;23:191–196. doi:10.1016/j.bjid.2019.05.006
13. Lipsky BA, Aragón‐Sánchez J, Diggle M, et al. IWGDF guidance on the diagnosis and management of foot infections in persons with diabetes. Diabetes Metab Res Rev. 2016;32:45–74.
14. Gosset P, Pichavant M, Sirard J-C, Bachirou K, Perez-cruz M Methods and pharmaceutical compositions for the treatment of acute exacerbations of chronic obstructive pulmonary disease. Google Patents; 2018.
15. Wang S-H, Sun Z-L, Guo Y-J, et al. Meticillin-resistant Staphylococcus aureus isolated from foot ulcers in diabetic patients in a Chinese care hospital: risk factors for infection and prevalence. J Med Microbiol. 2010;59(10):1219–1224. doi:10.1099/jmm.0.020537-0
16. Nelson EA, Backhouse MR, Bhogal MS, et al. Concordance in diabetic foot ulcer infection. BMJ Open. 2013;3(1).
17. Mathangi T, Prabhakaran P, Rayappan F, Tilton F. Isolation, molecular characterization and antibiogram of bacteria isolated from diabetic foot ulcers. Int J Curr Res Aca Rev. 2013;1(1):17–25.
18. Sastry AS, Bhat S. Essentials of Medical Microbiology. Jaypee Brothers, Medical Publishers Pvt. Limited; 2018.
19. CLSI. M100 Performance Standards for Susceptibility Testing. Clinical and Laboratory Standards Institute Wayne (PA); 2018.
20. Karmakar A, Jana D, Dutta K, Dua P, Ghosh C. Prevalence of panton-valentine leukocidin gene among community acquired Staphylococcus aureus: a real-time PCR study. J Pathog. 2018;2018.
21. Bhatta DR, Cavaco LM, Nath G, et al. Association of Panton Valentine Leukocidin (PVL) genes with methicillin resistant Staphylococcus aureus (MRSA) in Western Nepal: a matter of concern for community infections (a hospital based prospective study). BMC Infect Dis. 2016;16(1):199. doi:10.1186/s12879-016-1531-1
22. Karadağ FY, Saltoğlu N, Ö A, et al. Foot self-care in diabetes mellitus: evaluation of patient awareness. Prim Care Diabetes. 2019.
23. Verrone Quilici MT, Del Fiol F, Franzin VAE, Toledo MI. Risk factors for foot amputation in patients hospitalized for diabetic foot infection. J Diab Res. 2016;2016.
24. Abdul Qadir D, Ali SAAA, Mahmoud HM. Prevalence of bacterial types in Wagner grade three of diabetic foot ulcer. Iraqi J Sci. 2014;55(3B):1232–1235.
25. Anvarinejad M, Pouladfar G, Japoni A, et al. Isolation and antibiotic susceptibility of the microorganisms isolated from diabetic foot infections in Nemazee Hospital, Southern Iran. J Pathog. 2015;2015.
26. Akhi MT, Ghotaslou R, Memar MY, et al. Frequency of MRSA in diabetic foot infections. Int J Diab Dev Ctries. 2017;37(1):58–62. doi:10.1007/s13410-016-0492-7
27. Víquez-Molina G, Aragón-Sánchez J, Pérez-Corrales C, Murillo-Vargas C, López-Valverde ME, Lipsky BA. Virulence factor genes in Staphylococcus aureus isolated from diabetic foot soft tissue and bone infections. Int J Low Extrem Wounds. 2018;17(1):36–41. doi:10.1177/1534734618764237
28. Katz DE, Friedman ND, Ostrovski E, et al. Diabetic foot infection in hospitalized adults. J Infection Chemotherapy. 2016;22(3):167–173. doi:10.1016/j.jiac.2015.12.007
29. Dwedar R, Ismail DK, Abdulbaky A. Diabetic foot infection: microbiological causes with special reference to their antibiotic resistance pattern. Egyptian J Med Microbiol. 2015;38(3174):1–8.
30. Citron DM, Goldstein EJ, Merriam CV, Lipsky BA, Abramson MA. Bacteriology of moderate-to-severe diabetic foot infections and in vitro activity of antimicrobial agents. J Clin Microbiol. 2007;45(9):2819–2828. doi:10.1128/JCM.00551-07
31. Hatipoglu M, Mutluoglu M, Turhan V, et al. Causative pathogens and antibiotic resistance in diabetic foot infections: a prospective multi-center study. J Diab Complications. 2016;30(5):910–916. doi:10.1016/j.jdiacomp.2016.02.013
32. Kim TG, Moon SY, Park MS, et al. Factors affecting length of hospital stay and mortality in infected diabetic foot ulcers undergoing surgical drainage without major amputation. J Korean Med Sci. 2016;31(1):120–124. doi:10.3346/jkms.2016.31.1.120
33. Mottola C, Semedo-Lemsaddek T, Mendes JJ, et al. Molecular typing, virulence traits and antimicrobial resistance of diabetic foot staphylococci. J Biomed Sci. 2016;23(1):33. doi:10.1186/s12929-016-0250-7
34. Perim MC, Borges J, Celeste SRC, et al. Aerobic bacterial profile and antibiotic resistance in patients with diabetic foot infections. Rev Soc Bras Med Trop. 2015;48(5):546–554. doi:10.1590/0037-8682-0146-2015
35. Clinical, Institute LS. Performance Standards for Antimicrobial Susceptibility Testing; 17th Informational Supplement. Wayne, PA, USA: CLSI document M100‐S17 Clinical and Laboratory Standards Institute; 2007.
36. Baddour M, AbuElKheir M, Fatani A. Comparison of mecA polymerase chain reaction with phenotypic methods for the detection of methicillin-resistant Staphylococcus aureus. Curr Microbiol. 2007;55(6):473. doi:10.1007/s00284-007-9015-6
37. Deplano A, Vandendriessche S, Nonhoff C, Denis O. Genetic diversity among methicillin-resistant Staphylococcus aureus isolates carrying the mecC gene in Belgium. J Antimicrobial Chemotherapy. 2014;69(6):1457–1460. doi:10.1093/jac/dku020
38. Kavitha Y, Mohan S, Moinuddin SK. Bacteriological profile of diabetic foot infection with special reference to ESBL and MRSA in a coastal tertiary care teaching hospital. Indian J Microbiol Res. 2017;4(1):68–73.
39. Demir T, Coplu N, Esen B. Comparative analysis of phenotypic and genotypic detection of methicillin resistance among Staphylococcus aureus. Indian J Pathol Microbiol. 2016;59(3):314. doi:10.4103/0377-4929.188103
40. Stappers MH, Hagen F, Reimnitz P, Mouton JW, Meis JF, Gyssens IC. Direct molecular versus culture-based assessment of Gram-positive cocci in biopsies of patients with major abscesses and diabetic foot infections. European J Clin Microbiol Infectious Diseases. 2015;34(9):1885–1892. doi:10.1007/s10096-015-2428-4
41. Pobiega M, Myjak I, Pomorska-Wesolowska M, et al. Virulence potential of Staphylococcus aureus strains isolated from diabetic foot ulcers among patients from southern Poland. Curr Vasc Pharmacol. 2016;14(6):547–551. doi:10.2174/1570161114666160625083742
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

