Back to Journals » Infection and Drug Resistance » Volume 14
Antibacterial Effects of Disulfiram in Helicobacter pylori
Authors Kobatake T, Ogino K, Sakae H, Gotoh K, Watanabe A, Matsushita O, Okada H , Yokota K
Received 1 March 2021
Accepted for publication 12 April 2021
Published 12 May 2021 Volume 2021:14 Pages 1757—1764
DOI https://doi.org/10.2147/IDR.S299177
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Tomomi Kobatake,1 Keiki Ogino,2 Hiroyuki Sakae,3 Kazuyoshi Gotoh,4 Akari Watanabe,5 Osamu Matsushita,4 Hiroyuki Okada,3 Kenji Yokota1
1Graduate School of Health Science Okayama University, Okayama, 700-8558, Japan; 2Department of Environmental Medicine, Koch Medical School, Nankoku-shi, 783-8505, Japan; 3Department of Gastroenterology and Hepatology, Graduate School of Medicine Dentistry and Pharmaceutical Sciences, Okayama University, Okayama, 700-8558, Japan; 4Department of Bacteriology, Graduate School of Medicine Dentistry and Pharmaceutical Sciences, Okayama University, Okayama, 700-8558, Japan; 5Department of Oral Health Care and Rehabilitation, Institute of Biomedical Sciences, Tokushima University Graduate School, Tokushima, 770-8501, Japan
Correspondence: Kenji Yokota
Graduate School of Health Science Okayama University, Okayama, 700-8558, Japan
Tel/Fax +81-86-235-6846
Email [email protected]
Background: Helicobacter pylori infection poses a risk of the occurrence of gastrointestinal diseases, such as gastric cancer. Its incidence rate is significantly reduced by eradication, and thereby, eradication therapy is generally performed. Disulfiram is an oral prescription drug mainly used for the treatment of alcohol dependence. In recent years, reports have been made on its anticancer and antibacterial effects, and thus, it has recently become an interesting subject. This study aimed to examine the antibacterial activity of disulfiram, investigate the presence or absence of its antibacterial activity on H. pylori, and determine whether it could be a new bactericidal drug against drug-resistant H. pylori.
Materials and Methods: Drug-sensitive strains of H. pylori and amoxicillin-resistant, clarithromycin-resistant, and metronidazole-resistant strains were used, and a growth inhibition test of H. pylori using disulfiram was performed. Furthermore, the expression of urease, vacuolating cytotoxin A (VacA), and CagA, the virulence proteins of H. pylori, was quantitatively analyzed using the Western blotting method. In addition, for H. pylori used in this study, the 16SrDNA sequence, a ribosomal gene involved in protein production, was analyzed to examine the presence or absence of gene mutation.
Results: Disulfiram suppressed the growth of 7 out of 12 H. pylori strains at 1 μg/mL, and no correlation was observed between their susceptibility/resistance to current eradication antimicrobial drugs and disulfiram resistance. Disulfiram reduced the expression levels of urease, VacA, and CagA proteins. H. pylori, which showed resistance to disulfiram, tended to have fewer gene deletions/insertions in the 16S rDNA sequence; however, no specific mutation was detected.
Conclusion: Disulfiram has a bactericidal effect on H. pylori at low concentrations, suggesting that it can be used as a supplement for current H. pylori eradication drugs.
Keywords: disulfiram, Helicobacter pylori, urease, vacuolating toxin, CagA
Introduction
Helicobacter pylori is a Gram-negative spiral bacterium that inhabits the human gastric mucosa. H. pylori infection poses a risk of the occurrence of gastrointestinal diseases, such as peptic ulcer, gastric mucosa-associated lymphoid tissue lymphoma, and gastric cancer;1–4 however, its occurrence is significantly reduced by eradication.5,6 In Japan, eradication therapy using amoxicillin and clarithromycin is the primary treatment for H. pylori infection; however, in recent years, the eradication success rate is decreasing due to the increased incidence of drug-resistant H. pylori.7
Disulfiram is an oral administration drug marketed under the trade name Antabuse and used primarily indicated for the treatment of alcohol dependence. When disulfiram is orally ingested, one molecule of disulfiram is converted into two molecules of diethyldithiocarbamic acid in the gastrointestinal tract and blood and further metabolized into diethyldithiocarbamic acid methyl ester sulfoxide. When these metabolites react and bind to activation sites such as aldehyde dehydrogenase, alcohol metabolism is reduced, resulting in hangover symptoms, resulting in avoidance of alcohol inoculation.8 Disulfiram has been used as a therapeutic agent for alcohol dependence for many years; however, studies have re-focused on its anticancer9,10 and antibacterial activities in recent years.11,12
Therefore, this study focused on the antibacterial activity of disulfiram, investigated the presence or absence of its antibacterial activity against H. pylori, and investigated whether it could be a new bactericidal drug against drug-resistant H. pylori.
Materials and Methods
Strains
H. pylori strains were isolated from patients admitted to Okayama University Hospital with informed consent. Ethical approval to carry out the study was obtained a priori from the Okayama University Ethics Committee (Number 2034). H. pylori samples were collected in accordance with the 1964 Helsinki declaration and its later amendments. E-test was used to measure susceptibility to amoxicillin, clarithromycin, and metronidazole for antibacterial sensitivity. If the minimum inhibitory concentration (MIC) value of amoxicillin is ≥0.1 µg/mL; clarithromycin, ≥2 µg/mL; and metronidazole, ≥1 µg/mL, the strain is identified as resistant to each antibiotic. Then, drug-sensitive, amoxicillin-resistant, clarithromycin-resistant, and metronidazole-resistant strains were selected. Table 1 shows drug resistance values of the selected 12 strains.
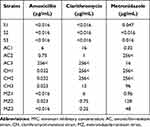 |
Table 1 MIC Values of H. pylori Strain Used |
H. pylori Growth Inhibition Test with Disulfiram
Whether disulfiram suppresses the growth of H. pylori was measured. About 20 mL of brain heart infusion (BHI) medium containing 5% horse serum was placed in a 50-mL liquid culture flask, and the medium was allowed to stand in a carbon dioxide (CO2) incubator overnight to saturate the liquid medium with CO2. Disulfiram (Sigma-Aldrich) was added to a CO2-saturated liquid medium to prepare a chemical medium with a disulfiram concentration of 10, 5, 1, and 0 µg/mL. As a pre-culture, H. pylori, which had been pure-cultured on BHI agar medium containing 7% horse defibrosis blood at 37°C for 4 days under microaerobic conditions, was suspended in a BHI solution containing 5% horse serum at OD600=1.0 (108 CFU/mL). H. pylori infused with 500 µL each suspended medium was inoculated and cultured by shaking at 37°C under microaerobic conditions for 4 days. Absorbance was measured at OD600 nm every day, and the bacterial growth was confirmed by increased absorbance. Bacterial solution was collected after shaking the culture, and morphological changes of the bacteria were observed with Gram staining. A portion of the cultured medium was applied to the horse blood BHI agar medium and cultured at 37°C in microaerobic conditions for 7 days to confirm its effects on H. pylori developmental ability.
Protein Expression Level Measurement
The amount of H. pylori was estimated from strain absorbance in which the growth of H. pylori was confirmed in the growth inhibition test. When OD600 = 0.5, the amount of bacteria is approximately 107 CFU/mL. Bacterial solutions were collected from media with disulfiram concentrations of 0 µg/mL and 1 µg/mL by centrifugation. Bacteria were re-suspended into distilled water with 1 mM of protease inhibitor phenyl-methyl-sulfonyl fluoride and disrupted by ultrasonic waves in ice for 10 min using a device (Bioruptor, Cosmobio). A sample was prepared by diluting the same amount as the SDS sample buffer and heating at 100°C for 10 min. About 10 of the prepared sample was added to 1 well, electrophoresed at 90 V for 2 h using the SDS-PAGE, and then transferred to a polyvinylidene fluoride (PVDF) membrane at 100 mA for 4 h. After the transfer, the PVDF membrane was blocked overnight with phosphate-buffered saline (PBS) containing 10% skim milk. After washing with PBS for 5 min and 3 times, urease (30 kDa) antibody (in house), CagA antibody (Funakoshi), or VacA antibody (Santa Cruz Biotechnology, Inc) diluted 1000-fold with 10% skim milk-added PBS were added as primary antibodies and shaken for 2 h; horseradish peroxidase-labeled antibody (Dako) diluted 1000-fold with 10% skim milk-added PBS was added as a secondary antibody and shaken for 1 h; and the color was developed by Pierce® Western Blotting Substrate (Thermo Fisher Scientific), and the band was detected and quantified by chemiluminescence using the Amersham Imager 600 (Cytiva).
Confirmation of Gene Mutation
DNA Extraction from the Bacteria
DNA was extracted from the strain according to the product protocol of QIAamp DNA Micro Kit (QIAGEN). The bacteria were suspended in a 1.5-mL Eppen tube with 500-µL of physiological saline and centrifuged at 15,000 rpm for 2 min, and the supernatant was discarded. The remaining precipitate was pulverized by tapping or vortexing, added with 300 μL of Nuclei Lysis Solution, and the mixture was suspended by pipetting, incubated at 80°C for 5 min, and cooled at room temperature. After adding 1.5 μL of RNase and mixing by inversion, the mixture was incubated at 37°C for 15 min and cooled at room temperature. Thereafter, 100 μL of protein precipitation solution was added, vortexed, and then cooled in ice for 5 min. Centrifugation was performed at 4°C and 15,000 rpm for 3 min, the supernatant was placed in a new Eppen tube, and 300 μL of isopropanol was added. Thereafter, the mixture was centrifuge at 4°C and 15,000 rpm for 2 min, the supernatant was discarded, and 300 μL of 70% ethanol was added. It was centrifuged again at 4°C and 15,000 rpm for 2 min, and the ethanol in the tube was carefully aspirated. After drying the tube at 37°C, 50 μL of DNA rehydration solution was added and incubated at 65°C for 60 min.
Polymerase Chain Reaction Detection and Sequence
The sequence of 16S rDNA, a ribosomal gene involved in protein production, was detected by polymerase chain reaction (PCR) using primers 16S-F and 16S-R. The sequence of each primer is shown in Table 2.
 |
Table 2 PCR Primers |
Using the DNA of the extracted bacteria as a template, PCR was performed in a total amount of 50 µL. TaKaRa Ex Taq (5 units/µL) of 0.5 µL; 10 × Ex Taq Buffer, 5.0 µL; dNTP mixture (2.5 mM each), 4.0 µL; template, 1.0 µL; primer pair, 1.0 µL; sterile distilled water, 38 µL were mixed. PCR conditions were 98°C for 1-min denaturation, 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s for 35 cycles. A 5-µL PCR product was electrophoresed on a 2% agarose gel for 30 min. The gel after migration was stained with ethidium bromide for 20 min and observed by ultraviolet irradiation using an electrophoresis gel imaging device Dolphin-View (Dolphin-View, Kurabo, Japan). The DNA sample amplified by PCR was purified using the ethanol precipitation method. The sequence was outsourced to the Hokkaido System Science Co., Ltd. Samples were sequenced using the PRISM3130 Genetic Analyzer (Applied Biosystems), and sequence data were analyzed with GENETYX software.
Statistical Analysis
Statistical analysis of H. pylori protein expression level measurement was performed using t-test. The P-value of <0.05 was considered statistically significant.
Results
H. pylori Growth Inhibition Test with Disulfiram
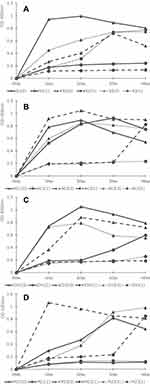
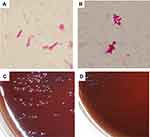
Whether disulfiram inhibits the growth of H. pylori was examined using a total of 12 strains. All strains underwent complete growth inhibition in the media with disulfiram concentrations of 10 µg/mL and 5 µg/mL. In the drug-sensitive (Figure 1A) and clarithromycin-resistant strains (Figure 1C), two strains were suppressed in growth in a medium with a disulfiram concentration of 1 µg/mL, and one strain grew from the third day of culture. In the metronidazole-resistant strain (Figure 1D), two strains were suppressed in growth in a medium with a disulfiram concentration of 1 µg/mL, and one strain grew from the 4th day of culture. Among the amoxicillin-resistant strains (Figure 1B), one strain underwent growth inhibition in a medium with a disulfiram concentration of 1 µg/mL, and one strain grew from the 4th day of culture, and another one strain grew from the 1st day of culture. No correlation was found between susceptibility/resistance to current disinfectants and disulfiram resistance. After collecting the bacterial solution after shaking the culture and observing the morphological changes of the bacteria using the Gram stain, the spiral shape was lost and the bacteria changed to a short rod-like (Figure 2A and B). In addition, when applied to BHI agar medium with 7% horse blood and cultured at 37°C for 7 days, the amount of colonies was smaller than the usual growth (Figure 2C and D).
H. pylori Pathogenic Protein Expression Level Measurement
Bacterial proteins were compared with a disulfiram concentration of 0 µg/mL and 1 µg/mL using one from each of the drug-sensitive, amoxicillin-resistant, clarithromycin-resistant, and metronidazole-resistant strain with growth confirmed in the growth inhibition test. The increase/decrease in urease, VacA, and CagA protein expressions levels in each bacterium was measured. In the drug-sensitive strain, protein expression levels of all three species decreased, and a significant difference was observed between urease and VacA (Figure 3A). Amoxicillin-resistant strains had significantly reduced protein expression levels in all three species (Figure 3B). In the clarithromycin-resistant strain, protein expression levels of all three species decreased, and a significant difference was observed in urease (Figure 3C). In the metronidazole-resistant strain, expression levels of urease and VacA decreased and that of CagA increased, and no significant difference was observed among the three species (Figure 3D).
Confirmation of Gene Mutation
For all 12 H. pylori strains used, the sequence of 16SrDNA, a ribosomal gene involved in protein production, was analyzed, and the presence or absence of gene mutation was examined. Table 3 shows the number of base substitutions and the number of deletions/insertions in the 16SrDNA sequence. Of these, susceptible strain 3 (S3), amoxicillin-resistant strain 1 (AC1), amoxicillin-resistant strain 2 (AC2), clarithromycin-resistant strain 1 (CH1), and metronidazole-resistant strain 2 (MZ2) were resistant to disulfiram in the growth suppression test. Strains resistant to disulfiram tended to have fewer base deletions/insertions.
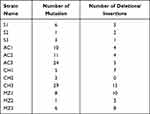 |
Table 3 16S rDNA Sequence |
Discussion
Eradication therapy using amoxicillin and clarithromycin is performed for H. pylori infection, a risk factor for the occurrence of gastrointestinal diseases such as gastric cancer. However, in recent years, a decrease in the eradication success rate due to an increase in drug-resistant strains has become a problem, and further increase in drug-resistant strains and the emergence of resistant strains to new antibiotics should be prevented.13
Disulfiram is an oral prescription drug mainly used as a therapeutic drug for alcohol dependence and is usually taken orally in 1–3 divided doses of 100–500 mg daily, and the maintenance dose is 100–200 mg.8 In this study, disulfiram suppressed the growth of 11 among 12 strains at a concentration of 1 µg/mL at 2 days of H. pylori culture and 7 at 4 days of culture. In addition, H. pylori acted on disulfiram showed poor growth of colonies in 7% horse blood BHI agar medium, and the cells lost their spiral shape and became short rod-like. Based on these facts, disulfiram is considered to exhibit an antibacterial effect against H. pylori at a low concentration regardless of the sensitivity and resistance to the current H. pylori eradication drug.
Urease, VacA, and CagA are important proteins with pathogenic effect in the stomach. H. pylori, which neutralized gastric acid with urease, secretes VacA, a cell-vacuolating toxin, destroys host cells, and secretes CagA protein to promote carcinogenesis of host cells.14 In this study, the expression levels of urease, VacA, and CagA proteins decreased in H. pylori infection treated with disulfiram. Based on the results of the growth inhibition test, disulfiram is considered to exhibit a concentration-dependent bactericidal action by inhibiting protein synthesis of H. pylori.
16SrDNA is a ribosomal gene involved in protein production, and this sequence of approximately 1500 bases is used for bacterial phylogeny. The 16SrDNA sequence is abundantly registered in public databases and is considered suitable for examining sequences specific to a particular bacterial species/group.15,16 When 16SrDNA mutations were confirmed in all 12 H. pylori strains used in this study, strains showing disulfiram resistance tended to possess fewer base deletions/insertions but were susceptible/resistant to disulfiram. No specific mutations due to differences were detected. Most of these deletions/insertions were found in domain 2 (V4 region) of 16SrDNA, and we cannot find that this fact is important or associate with protein production. For clearer features, the number of samples should be increased, and further study conducted.
Conclusion
Although the genetic characteristics on the effectiveness of disulfiram against H. pylori can be further investigated, it exhibited a bactericidal effect on H. pylori at a lower concentration than when used as a supplemental drug. Therefore, disulfiram could be used as an adjunct to the current H. pylori eradication drug.
Abbreviations
AC, amoxicillin; CH, clarithromycin; MZ, metronidazole; BHI, brain heart infusion; MIC, minimum inhibitory concentration; PCR, polymerase chain reaction.
Acknowledgment
This research was supported by Research Grant No19HA1007 of Ministry of Health, Labor and Welfare Science.
Disclosure
The authors report no conflicts of interest in this work.
References
1. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Infection with Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994;61:177–240.
2. Blaser MJ. The bacteria behind ulcers. Sci Am. 1996;274(2):104–107. doi:10.1038/scientificamerican0296-104
3. Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784–789. doi:10.1056/NEJMoa001999
4. Graham DY. Helicobacter pylori infection is the primary cause of gastric cancer. J Gastroenterol. 2000;35:90–97.
5. Lee YC, Chiang TH, Chou CK, et al. Association between Helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology. 2016;150(5):1113–1124. doi:10.1053/j.gastro.2016.01.028
6. Rokkas T, Rokka A, Portincasa P. A systematic review and meta-analysis of the role of Helicobacter pylori eradication in preventing gastric cancer. Ann Gastroenterol. 2017;30(4):414–423. doi:10.20524/aog.2017.0144
7. Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization Regions. Gastroenterology. 2018;155(5):1372–1382. doi:10.1053/j.gastro.2018.07.007
8. Franck J, Jayaram-Lindström N. Pharmacotherapy for alcohol dependence: status of current treatments. Curr Opin Neurobiol. 2013;23(4):692–699. doi:10.1016/j.conb.2013.05.005
9. Skrott Z, Mistrik M, Andersen KK, et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature. 2017;552(7684):194–199. doi:10.1038/nature25016
10. Viola-Rhenals M, Patel KR, Jaimes-Santamaria L, Wu G, Liu J, Dou QP. Recent advances in antabuse (disulfiram): the importance of its metal-binding ability to its anticancer activity. Curr Med Chem. 2018;25(4):506–524. doi:10.2174/0929867324666171023161121
11. Sheppard JG, Frazier KR, Saralkar P, Hossain MF, Geldenhuys WJ, Long TE. Disulfiram-based disulfides as narrow-spectrum antibacterial agents. Bioorg Med Chem Lett. 2018;28(8):1298–1302. doi:10.1016/j.bmcl.2018.03.023
12. Frazier KR, Moore JA, Long TE. Antibacterial activity of disulfiram and its metabolites. J Appl Microbiol. 2019;126(1):79–86. doi:10.1111/jam.14094
13. Alba C, Blanco A, Alarcón T. Antibiotic resistance in Helicobacter pylori. Curr Opin Infect Dis. 2017;30(5):489–497. doi:10.1097/QCO.0000000000000396
14. Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7(11):629–641. doi:10.1038/nrgastro.2010.154
15. Christensen JJ, Andresen K, Justesen T, Kemp M. Ribosomal DNA sequencing: experiences from use in the Danish National Reference Laboratory for Identification of Bacteria. APMIS. 2005;113(9):621–628. doi:10.1111/j.1600-0463.2005.apm_224.x
16. Woo PCY, Lau SKP, Teng JLL, Tse H, Yuen K-Y. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 2008;14(10):908–934.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.