Back to Journals » Journal of Inflammation Research » Volume 16
An Integrated Analysis Reveals Ciliary Abnormalities in Antrochoanal Polyps
Authors Zi X, Peng Y, Zang Y, Chen S, Li M, Yu K, Liang X, Jin P , Wang D , Shi L
Received 22 November 2022
Accepted for publication 26 January 2023
Published 14 February 2023 Volume 2023:16 Pages 605—615
DOI https://doi.org/10.2147/JIR.S398371
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Ning Quan
Xiaoxue Zi,1,* Yang Peng,2,* Yiran Zang,1 Shiying Chen,2 Mengshi Li,3 Kena Yu,1 Xu Liang,1 Peng Jin,1 Deyun Wang,4 Li Shi1
1Department of Otolaryngology, The Second Hospital of Shandong University, Jinan, People’s Republic of China; 2State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou Medical University, Guangzhou, People’s Republic of China; 3State Key Laboratory of Respiratory Disease, Department of Respiratory Pathology, Guangzhou Institute of Respiratory Health, National Clinical Research Center for Respiratory Disease, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, People’s Republic of China; 4Department of Otolaryngology, National University of Singapore, National University Health System, Singapore
*These authors contributed equally to this work
Correspondence: Peng Jin, Department of Otolaryngology, The Second Hospital of Shandong University, 247 Beiyuan Avenue, Jinan, Shandong, 250033, People’s Republic of China, Tel +86 531 85875317, Fax +86 531 88962544, Email [email protected]
Objective: The mechanisms underlying the antrochoanal polyps (ACPs) remained unclear. We aimed to identify the differentially expressed genes (DEGs) profile, the cilia-related genes expression levels and the morphological characteristics of ciliated cells in patients with ACPs.
Methods: We obtained ACPs biopsy samples from 28 patients and uncinate process from 27 healthy controls. Whole-transcriptome RNA sequencing, immunofluorescence staining, quantitative polymerase chain reaction, and scanning electron microscopy were performed.
Results: 3739 DEGs were detected between ACPs and controls, and Gene Ontology analysis on these DEGs implicated cilium assembly, cilium motility, cilia component, cilia function, inflammatory response and immune system process were included in ACPs pathogenesis. Gene set enrichment analysis implicated sets of genes regulated in processes associated with cilium organization, cilium morphogenesis, cilium movement, axoneme assembly, axonemal dynein complex assembly and cell projection assembly. The expression levels of cilia-related genes (FOXJ1, DNAI1, DNAH9, RSPH1, RSPH9 and RSPH4A) were validated by quantitative polymerase chain reaction (Fold change > 2, P< 0.05) and FOXJ1 was positive correlated with DNAI1, DNAH9, RSPH4, RSPH1, RSPH9, DNAH5, DNALI1 in ACPs (all P < 0.05). Based on our semi-quantitative scoring system, median scores of α-Tubulin, DNAI1 and RSPH4A were significantly higher in ACPs than in controls. In addition, loss of ciliated cells and a shorter cilia pattern were further confirmed by immunofluorescence staining and scanning electron microscopy in ACPs.
Conclusion: The aberrant expression of cilia-related genes and ciliary structural impairment are an important pathological phenomenon in ACPs, and our findings may provide novel insights into understanding the mysterious mechanisms underlying ACPs.
Keywords: antrochoanal polyps, ciliary abnormalities, transcriptome RNA sequencing, uncinate process
Introduction
The antrochoanal polyps (ACPs) are occurred more frequently in children and young adults and are believed to be caused by mucous gland obstruction in maxillary sinus mucosa after chronic inflammation.1,2 Clinically, an ACP appears as a bright, grey or pinkish mass in the middle meatus and nasal cavity protruding posteriorly to the choana. Although endoscopic sinus surgery is commonly accepted as a treatment for ACPs, the recurrence rate of ACPs can be up to 21% after surgery.3,4 These problems arise partly because the underlying pathogenic mechanisms of ACPs are poorly understood.5
MCC provides the first line of defense for airway by removing foreign particles through the coordination between secretory cells and ciliated cells.6 The potential consequence of MCC impairment is retention of inhaled substances (micro-organisms, cigarette smoke, air pollutants, etc.), leading to chronic inflammation in respiratory tract. It has been reported that MCC abnormality of the airway epithelium has been closely associated with the pathogenesis of several airway inflammatory disease (eg, allergic rhinitis, nasal polyps, asthma and idiopathic pulmonary fibrosis).7–11 However, it remains elusive whether the MCC abnormalities are common pathologic characteristics in ACP patients, and further studies to detect the expression levels of MCC related markers as well as to characterize the morphological features of ciliated cells in ACP tissues are warranted.
RNA sequencing (RNA-seq) is a powerful and unbiased mean to explore candidate transcripts.11 In this study, we analyzed the profiles of gene expression in polyp tissues from ACPs patients and uncinate process from control subjects, as well as further delineated a transcriptome picture and identified key pathways that are dysregulated in ACPs. We also conducted polymerase chain reaction, immunofluorescence (IF) staining and scanning electron microscopy (SEM) to confirm the ciliary abnormality revealed by RNA-seq.
Materials and Methods
Patient Recruitment
We recruited patients with biopsy-proven unilateral ACPs (n = 28), whose polyps were in the maxillary sinus, growing through the sinus ostium and posterior nasal cavity, and extending into the choana and nasopharynx. All patients performed preoperative computed tomography (CT) scans before surgery and subsequently underwent endoscopic nasal surgery in The Second Hospital of Shandong University. Uncinate process (n = 27) biopsies were obtained from subjects with simple maxillary sinus cyst and symptomatic nasal septal deviation requiring surgery, which were defined as controls (Table 1). No study participant had a physician’s primary diagnosis of asthma, primary ciliary dyskinesia, or cystic fibrosis. Atopy was evaluated with serum allergen testing and a positive serum specific IgE (≥0.35 kU/L, by CAP Pharmacia, Uppsala, Sweden) to at least one or more common local allergens was confirmed. Due to the limited sizes of the tissue, not all specimens were used for each analysis. This study complies with the Declaration of Helsinki. The written informed forms from all participants in this study was obtained. Approval for this study was obtained from the institutional review board (IRB) of the Second Hospital of Shandong University, China [China approval number: KYLL-2018 (KJ) P-0028].
 |
Table 1 Summary Characteristics of Study Participants |
RNA Sequencing, Data Processing and Bioinformatics Analysis of the RNA-Seq
Fresh tissues were pre-treated with RNAlaterTM (Thermo Fisher Scientific, Waltham, MA, USA), the RNA was extracted by standard extraction method, RNA of each sample were used as input material and sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated. Raw data was qualified controlled and gene expression level was quantified. Details steps are shown in the Supplementary Text.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from ACPs and control tissues. Details steps are shown in the Supplementary Text. The mRNA expression levels of 9 cilia-related genes (FOXJ1, DNAI1, DNAH9, RSPH1, RSPH9, RSPH4A, CP110, DNAH5, DNALI1) and 8 inflammatory factors (CCL7, CCL18, CCL20, CXCL6, CXCL8, IL17REL, IFNG and IL13RA2) were validated with qRT-PCR. The relative expression was calculated by using the 2−ΔΔCt methods normalized against the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The details of primer sequences, Fold change, and P values are presented in Tables E8 and E9.
Hematoxylin-Eosin (HE) and IF Assays
Paraffin tissue sections were processed to perform HE and IF assay as previously reported.12 The sections were incubated with primary antibodies overnight at 4 °C followed by incubation with Alexa Fluor conjugated secondary antibodies (Life Technologies, Carlsbad, CA, USA) at 37 °C for 1 hour. Cellular nuclei were visualized by staining with 4’,6-diamidino-2-phenylindole (Life Technologies). Images were acquired with fluorescence microscope (Olympus IX51, Tokyo, Japan). Detail of antibodies and steps for IF are shown in the Supplementary Text.
Evaluation of HE and IF Assays
All cases were assessed in a blinded fashion by independent researchers. Eosinophil infiltration was evaluated based on HE staining. Eosinophils were enumerated at five high-power fields (HPF, 400 × magnification) and denoted by higher than 10 eosinophils per HPF.13
Ciliary distribution and ciliary length were evaluated by IF staining with α-tubulin in five areas. For distribution, each area was assigned as score 0 (normal ciliary distribution was defined if the field contains >70% of epithelial areas with ciliary staining) or score 1 (ciliary shedding was defined if the field contains ≤ 70% of epithelial areas with ciliary staining).14 Ciliary length was measured by Image J software and 20 measurements for each area were recorded. The mean values of ciliary length were calculated before analysis.
For aberrant expression of ciliary ultrastructural markers (DNAI1 and RSPH4A), each area was assigned a score of 0 (normal expression) or 1 (aberrant expression). For which 0 point was assigned if the field contains >70% of ciliary distribution and 1 point was scored if the field contains ≤ 70% of ciliary distribution.14
The total fluorescence intensity (TFI, presented in arbitrary units) measurements were performed to evaluate FOXJ1, mucin 5AC (MUC5AC) and keratin 5 (KRT5) expressions. The positively stained area and mean fluorescence intensity (MFI) of each marker were recorded using Image J software. The TFI was calculated as the product of the positively stained area and the MFI.
SEM
Nasal tissues were fixed in 2.5% glutaraldehyde at 4 °C for overnight incubation. Samples were osmicated with 1% osmium tetroxide for 1 hour followed by dehydration with gradually increasing ethanol concentrations. Dried samples were mounted onto aluminum stubs and the surface was sputter-coated with gold for visualization with a FEI Quanta 250 FEG scanning electron microscope. Representative photomicrographs were taken at various angles so that any error in assessment was minimized because of the tilt of the specimen or other processing artifacts.
Statistical Analysis
Statistical analyses were conducted with GraphPad Prism 7 (GraphPad Inc., USA). For RNA sequence data, differentially expressed genes (DEGs) were identified based on the Fold change (cutoff: 2.0; >2.0 as upregulated and <-2 as downregulated) and Q values (cutoff: 0.05). For GO and KEGG analysis, the significant terms were retained if the Q value was less than 0.05. Mann–Whitney 2-sided nonparametric test was applied to compare the age, length of cilia, mRNA expression levels in qRT-PCR and immunofluorescence staining analysis. The correlation analysis was performed by Spearman correlation analysis. The associations between gender and allergen sensitization were analyzed by χ2 test. P < 0.05 was deemed statistically significant for all analyses.
Results
Subject Characteristics
The demographic data of ACPs patients and control subjects enrolled in this study were listed in Table 1. ACPs patients were male predominant, and the atopy statue was diagnosed in 0% (0/27) control subjects and 3.6% (1/28) ACPs patients. None of recruited participants had physician-diagnosed or self-reported asthma. The H&E staining showed that 7.7% (1/13) of ACPs tissues were eosinophilic.
Global Transcriptomic Profiling Reveals Ciliary Abnormalities in ACPs
A total of 3739 DEGs, including 2377 upregulated DEGs and 1362 downregulated DEGs, were identified between ACPs tissues and uncinate process tissues by RNA-seq (Fold change > 2 or < - 2; adj. P < 0.05). The volcano plot and heat map of DEGs were respectively shown in (Figure 1A and B), and the details of the top 20 upregulated and downregulated genes were summarized in Supplemental Tables E1. We found that cilia-related genes including FOXJ1, DNAI1, DNALI1, DNAH5, DNAH9, RSPH1, RSPH9, RSPH4A were significantly up-regulated in ACPs (Table E9).
GO and KEGG Analyses of the DEGs Were Performed
The top 20 GO categories [including biological processes (BP), cellular component (CC) and molecular function (MF)] and KEGG pathways were respectively listed in (Figure 1C–F) and Supplemental Tables E2–E5. Of the top 20 BP, 4 items were consistently related to cilium assembly and motility. Of the top 20 CC, 5 items were consistently related to cilia component. Of the top 20 MF, 4 items were consistently related to cilia function.
Furthermore, the enrichment analysis GSEA parameters details were shown in Table E6. Representative candidate enriched gene sets revealed cilium organization, cilium morphogenesis, cilium movement, axoneme assembly, axonemal dynein complex assembly, cell projection assembly, etc. (Figure 1G–L), Supplemental Table E7).
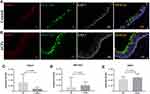
qRT-PCR Assay Validates Ciliary Abnormalities and Abnormal-Regulated Inflammation-Related Genes in ACPs
To validate the above-mentioned findings regarding cilia-related and inflammation-related DEGs expressions, we analyzed the expression levels of the 9 common cilia-related genes and 8 significantly abnormal-regulated inflammation-related genes in GO and analysis KEGG pathway analyses by qRT-PCR (Supplemental Tables E8 and E9). Except for DNALI1 (mean Fold change: 0.533, P = 0.718) and DNAH5 (mean Fold change: 0.822, P = 0.675), findings of qRT-PCR assay of FOXJ1, DNAI1, DNAH9, RSPH1, RSPH9 and RSPH4A were globally in accordance with the RNA-seq (Fold change > 2, P < 0.05). Besides, no significant difference of CP110 mRNA expression level was found between ACPs and uncinate process tissues (Figure 2). We further performed the correlation analysis and found the mRNA expression level of ciliogenesis marker (FOXJ1) was respectively positive correlated with ciliary ultrastructural markers (DNAI1, DNAH9, RSPH4, RSPH1, RSPH9, DNAH5, DNALI1) in biopsy tissues of ACPs (all P < 0.05) (Figure 2).
 |
Figure 2 Comparison of the expression levels of cilia-related genes in nasal mucosa between patients with ACPs (n = 18) and controls (n = 20) and their correlations in ACPs (n = 18). Shown in the figure are the expression levels of FOXJ1 (A), DNAI1 (B), DNAH9 (C), RSPH4A (D), RSPH1 (E), RSPH9 (F), CP110 (G), DNAH5 (H) and DNALI1 (I) in patients with ACPs and controls. The correlations between FOXJ1 and cilia-related genes expression levels are demonstrated in (Figure 4) (J–Q). Data were analyzed by Mann–Whitney 2-sided nonparametric test. Median values with range are indicated. The correlation analysis was performed by Spearman correlation analysis. P < 0.05 was statistically significant. FOXJ1, forkhead box J1; DNAI1, dynein axonemal intermediate chain 1; DNAH9, dynein axonemal heavy chain 9; RSPH1, radial spoke head 1 homolog; RSPH9, radial spoke head 9 homolog; RSPH4A, radial spoke head protein 4A; CP110, centrosomal protein 110; DNAH5, dynein axonemal heavy chain 5; DNALI1, dynein axonemal light intermediate chain 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. Abbreviations: ACPs, antrochoanal polyps; qRT-PCR, quantitative reverse transcription polymerase chain reaction. |
In addition, we found significantly up-regulated expression of CCL7, CCL18, CCL20, CXCL6, CXCL8, IL17REL and down-regulated expression of IFNG and IL13RA2 (Supplemental Figure E1). Next, we also analyzed the correlations between the mRNA expression of ciliogenesis marker (FOXJ1) and these inflammatory DEGs, and we found the expression level of FOXJ1 was significantly negative correlated with IL13RAL2 (Supplemental Figure E2).
Aberrant Protein Expression Level of Cilia Distribution, Ciliary Ultrastructural and Ciliogenesis Markers in ACPs
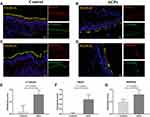
We further examined the distribution patterns of ciliated cells, the expression of ciliary ultrastructural (α-Tubulin, DNAI1 and RSPH4A) and ciliogenesis related marker (FOXJ1) in the nasal epithelium by performing IF staining and measured the length of cilia in both ACPs and control tissues.
Based on our semi-quantitative scoring system, the median (the 1st and 3rd quartile) scores for ciliary structure markers (α-Tubulin, DNAI1 and RSPH4A) were significantly increased in ACPs as compared to those of controls: α-Tubulin [0.8 (0.6, 1) vs 0.1 (0, 0.35)], DNAI1 [0.6 (0.2, 0.8) vs 0 (0, 0.05)] and RSPH4A [0.8 (0.5, 1) vs 0.5 (0.35, 0.6), P <0.001], respectively (all P < 0.05) (Figure 3).
Similar to control, FOXJ1 in ACPs was stained within the nucleus of both ciliated and nonciliated epithelial cells. Compared with control subjects, the TFI (arbitrary units) for FOXJ1 staining was markedly weaker in ACPs [8.25×106 (4.93×106, 1.57×106) vs 2.40×106 (8.18×106, 3.72×106), P < 0.005] (Figure 4).
Besides, the TFI of MUC5AC and KRT5 expression was non-significantly greater in ACPs than in control [5.05×106 (2.52×106, 8.31×106) vs 3,391,055 (1.03×106, 1.23×107), P = 0.443; 2.51×107 (2.22×107, 3.17×107) vs 2.20×107 (1.59×107, 2.68×107), P = 0.208] (Figure 4).
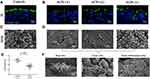
Aberrant Cilia Morphology of ACPs Under SEM
The normal ciliary morphology in control epithelium was relatively uniform distribution (Figure 5A and C) while abnormal ciliary morphology was observed in ACPs samples, including the loss of ciliated cells and a shorter cilia pattern (Figure 5B, D and F). In addition, the mean of cilia length being evaluated by IF staining was significantly shorter in ACPs than in controls (Figure 5E).
Discussion
ACPs are benign polypoid lesions with approximately 4–6% of all polyps in general population and about 35% of polyps in pediatric cases.1,4,15,16 However, the etiology and pathogenesis of ACPs still remain obscure. In this study, we performed transcriptomic sequencing followed by comparisons of ACPs and uncinate process. Results showed the global gene expression patterns differed between tissues of ACPs as compared with control subjects. Building on the RNA sequencing results and our previous research, we further revealed 9 significant aberrant cilia-related genes expression and ciliary impairment are both common characteristics of ACPs, which might provide important complementary insights into this disease.
Increasing amounts of studies identified various cytokines and chemokines involved in the polypogenesis of ACPs17,18 that help to understand the pathogenesis of ACPs.19 Our team have also previously demonstrated that ACPs is more likely associated with none type 2-driven inflammation and epithelial remodeling.5 In this study, KEGG pathway also analyses revealed chemokine signaling pathway, inflammatory mediator regulation of TRP channels and IL-17 signaling pathway, which reportedly correlates with chronic inflammation in ACPs. We further verified CCL7, CCL18, CCL20, CXCL6, CXCL8 and IL17REL were significantly up-regulated while IFNG and IL13RA2 were down-regulated in ACPs than controls. In addition, mRNA expression of ciliogenesis marker FOXJ1 and IL13RAL2 were negatively correlated in ACPs samples. According to the literature, these pathways play important roles in the pathophysiological alterations of several airway chronic inflammation diseases.19–22 However, the chronic inflammation of ACPs is still remaining obscure. As important components of the innate immune system, epithelium, mucous layer and mucociliary transport play important role during the occurrence and development of ACPs.13,23 Our GO analysis, including BP, CC and MF GO analysis, revealed genes associated with ciliary assembly, ciliary motility and ciliary function and inflammatory response. In addition, we further performed the enrichment analysis using GSEA to exert the biological functions in ACPs compared with control. Gene sets associated with cilium organization, cilium morphogenesis, cilium movement, axoneme assembly, axonemal dynein complex assembly, cell projection assembly was implicated, which partially echoed our findings by GO and KEGG. In brief, the ciliary abnormalities findings in our study may help to better understand the chronic inflammation in nasal mucosa of ACPs.
Physiologically, the proper structure of cilia is the basis of normal ciliary motility. We also found that the expression levels of basal cell markers KRT5 as well as goblet cell marker mucin 5AC were comparable between ACPs and controls. In addition, ciliogenesis is crucial to the formation of axonemes where ciliary ultrastructural proteins are assembled. Abnormal expression of ciliary ultrastructural markers has been implicated in chronic lower airway inflammatory diseases that have previously been linked to genetic defects, such as DNAH5, DNALI1, GAS8 (a component of the nexin-dynein regulatory complex), RSPH4A, RSPH9 and RSPH1.24,25 According to literatures, secondary ciliary dyskinesia was found in patients with chronic rhinosinusitis,7,8,26,27 bronchiectasis28 and chronic obstructive pulmonary disease.29 The abnormal ciliary ultrastructure consisted mainly of compound cilia, microtubular and dynein arm defects.27 Our published study has demonstrated the abnormal-regulation of ciliogenesis markers (CP110 and FOXJ1) and overly dense and lengthened cilia, greater prevalence of absence of DNAH5, RSPH1, RSPH4A and RSPH9 expressions, and the significantly reduced ciliary beat frequency have also been observed in NPs in the aberrantly remodeled epithelium of NPs.7,8,26 However, the characteristics of ciliary impairment in ACPs remain not fully explored. Therefore, according to the result of our sequencing analysis, we evaluated the expression of 9 common cilia-related genes and 2 ciliogenesis markers by qPCR analysis and IF staining. Consistent with the sequencing analysis findings, we found the abnormal mRNA expression of the cilia-related genes and the mRNA expression level of ciliogenesis marker (FOXJ1) was respectively positive correlated with ciliary ultrastructural markers (DNAI1, DNAH9, RSPH4, RSPH1, RSPH9, DNAH5, DNALI1) in biopsy tissues of ACPs. IF staining results further revealed aberrant protein expression of cilia distribution, ciliary ultrastructural and ciliogenesis markers in ACPs. So based on the results of our previous and present studies, we believe that ciliary abnormalities are common characteristics of NPs in general. The potential consequence of ciliary abnormalities is retention of inhaled substances, leading to chronic inflammation in airway and contributing to serious disorders in nasal cavity and sinuses, which might be an important factor of the ACPs recurrence. As reported in recent systematic review, despite continuous improvement in the surgical method and strive to remove the polyps as completely as possible, the recurrence rate of ACPs in children is still as high as 11%, even reaching to 15% reported.4,15,30 Therefore, further research elucidating the association between altered ciliary abnormalities and ACPs formation is needed.
In our study, the SEM image showed relatively uniform distribution in control epithelium, while the loss of cilia and shorter cilia pattern were found in ACPs epithelium. Interestingly, APCs has up-regulation of ciliogenesis marker (FOXJ1) and ciliary ultrastructural transcriptome but short cilia with a reduced number. On the one hand, the mRNA expression levels may cannot reliably reflect ciliary defects associated with aberrant FOXJ1 expression. On the other hand, the short cilia morphology and their absence on the epithelial cells may contribute to negative feedback of genetic expression. Further functional validation of the cilia-associated genes is warranted to elucidate the mechanisms underlying ACPs.
Our study has some limitations. First, the sample size (n=3) of SEM was too small in our study. However, we performed the SEM and IF in the 3 same samples [ACP(a), ACP(b) and ACP(c)] for compare the morphological aberration of ciliated epithelium between two methods. The results showed that IF staining could also reflect the aberrant ciliary distribution and length observed by SEM (Figure E5). Second, we only evaluated the significantly abnormal-regulated inflammation-related genes in GO and analysis KEGG pathway analyses, and in-depth investigation on the mechanism underlying the correlation between ciliary abnormality and inflammation are warranted.
In conclusion, our finding indicated that the aberrant expression of cilia-related genes and ciliary structural impairment are an important pathological phenomenon in ACPs, and may provide novel insights into understanding the mysterious mechanisms underlying ACPs.
Acknowledgment
The authors thank Dr Qiuliang Zhao, Dr Hongping Zhang, Dr Jiandong Zhao, Dr Tao Jiang for kindly supporting with the nasal specimens for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; they took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the report has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was supported by National Natural Science Foundation of China (81800885, 82101195 and 81873692), the Nature Science Foundation of Shandong Province (ZR2018PH021) and China Biodiversity Conservation and Green Development Foundation (cbcgdf-R122KQ001).
Disclosure
The authors declare no conflicts of interest.
References
1. Yaman H, Yilmaz S, Karali E, et al. Evaluation and management of antrochoanal polyps. Clin Exp Otorhinolaryngol. 2010;3(2):110–114. doi:10.3342/ceo.2010.3.2.110
2. Frosini P, Picarella G, De Campora E. Antrochoanal polyp: analysis of 200 cases. Acta Otorhinolaryngol Ital. 2009;29(1):21–26.
3. Chaiyasate S, Roongrotwattanasiri K, Patumanond J, et al. Antrochoanal polyps: how long should follow-up be after surgery? Int J Otolaryngol. 2015;2015:297417. doi:10.1155/2015/297417
4. Galluzzi F, Pignataro L, Maddalone M, et al. Recurrences of surgery for antrochoanal polyps in children: a systematic review. Int J Pediatr Otorhinolaryngol. 2018;106:26–30. doi:10.1016/j.ijporl.2017.12.035
5. Jin P, Zi X, Charn TC, et al. Histopathological features of antrochoanal polyps in Chinese patients. Rhinology. 2018;56(4):378–385. doi:10.4193/Rhin18.057
6. Jiao J, Zhang L. Influence of intranasal drugs on human nasal mucociliary clearance and ciliary beat frequency. Allergy Asthma Immunol Res. 2019;11(3):306–319. doi:10.4168/aair.2019.11.3.306
7. Zi XX, Guan WJ, Peng Y, et al. An integrated analysis of radial spoke head and outer dynein arm protein defects and ciliogenesis abnormality in nasal polyps. Front Genet. 2019;10:1083. doi:10.3389/fgene.2019.01083
8. Li YY, Li CW, Chao SS, et al. Impairment of cilia architecture and ciliogenesis in hyperplastic nasal epithelium from nasal polyps. J Allergy Clin Immunol. 2014;134(6):1282–1292. doi:10.1016/j.jaci.2014.07.038
9. Peng Y, Zi XX, Tian TF, et al. Whole-transcriptome sequencing reveals heightened inflammation and defective host defence responses in chronic rhinosinusitis with nasal polyps. Eur Respir J. 2019;54(5):1900732. doi:10.1183/13993003.00732-2019
10. Thomas B, Rutman A, Hirst RA, et al. Ciliary dysfunction and ultrastructural abnormalities are features of severe asthma. J Allergy Clin Immunol. 2010;126(4):722–729. doi:10.1016/j.jaci.2010.05.046
11. Peng Y, Wang ZN, Xu AR, et al. Mucus hypersecretion and ciliary impairment in conducting airway contribute to alveolar mucus plugging in idiopathic pulmonary fibrosis. Front Cell Dev Biol. 2021;9:810842. doi:10.3389/fcell.2021.810842
12. Peng Y, Guan WJ, Tan KS, et al. Aberrant localization of FOXJ1 correlates with the disease severity and comorbidities in patients with nasal polyps. Allergy Asthma Clin Immunol. 2018;14:71. doi:10.1186/s13223-018-0296-z
13. Fokkens WJ, Lund VJ, Hopkins C, et al. European position paper on rhinosinusitis and nasal polyps 2020. Rhinology. 2020;58(Suppl S29):1–464. doi:10.4193/Rhin20.401
14. Peng Y, Guan WJ, Zhu ZC, et al. Microarray assay reveals ciliary abnormalities of the allergic nasal mucosa. Am J Rhinol Allergy. 2020;34(1):50–58. doi:10.1177/1945892419871795
15. Zheng H, Tang L, Song B, et al. Inflammatory patterns of antrochoanal polyps in the pediatric age group. Allergy Asthma Clin Immunol. 2019;15:39. doi:10.1186/s13223-019-0352-3
16. Choudhury N, Hariri A, Saleh H. Endoscopic management of antrochoanal polyps: a single UK centre’s experience. Eur Arch Otorhinolaryngol. 2015;272(9):2305–2311. doi:10.1007/s00405-014-3163-7
17. Wang BF, Cao PP, Long XB, et al. Distinct mucosal immunopathologic profiles in atopic and nonatopic chronic rhinosinusitis without nasal polyps in Central China. Int Forum Allergy Rhinol. 2016;6(10):1013–1019. doi:10.1002/alr.21799
18. Konig K, Klemens C, Haack M, et al. Cytokine patterns in nasal secretion of non-atopic patients distinguish between chronic rhinosinusitis with or without nasal polys. Allergy Asthma Clin Immunol. 2016;12:19. doi:10.1186/s13223-016-0123-3
19. Wen W, Liu W, Zhang L, et al. Increased neutrophilia in nasal polyps reduces the response to oral corticosteroid therapy. J Allergy Clin Immunol. 2012;129(6):1522–1528. doi:10.1016/j.jaci.2012.01.079
20. Khorasanizadeh M, Eskian M, Gelfand EW, et al. Mitogen-activated protein kinases as therapeutic targets for asthma. Pharmacol Ther. 2017;174:112–126. doi:10.1016/j.pharmthera.2017.02.024
21. Bonvini SJ, Belvisi MG. Cough and airway disease: the role of ion channels. Pulm Pharmacol Ther. 2017;47:21–28. doi:10.1016/j.pupt.2017.06.009
22. Lee YK, Mukasa R, Hatton RD, et al. Developmental plasticity of Th17 and Treg cells. Curr Opin Immunol. 2009;21(3):274–280. doi:10.1016/j.coi.2009.05.021
23. Duan C, Li CW, Zhao L, et al. Differential expression patterns of EGF, EGFR, and ERBB4 in Nasal polyp epithelium. PLoS One. 2016;11(6):e156949. doi:10.1371/journal.pone.0156949
24. Antony D, Becker-Heck A, Zariwala MA, et al. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum Mutat. 2013;34(3):462–472. doi:10.1002/humu.22261
25. Shoemark A, Frost E, Dixon M, et al. Accuracy of immunofluorescence in the diagnosis of primary ciliary dyskinesia. Am J Respir Crit Care Med. 2017;196(1):94–101. doi:10.1164/rccm.201607-1351OC
26. Qiu Q, Peng Y, Zhu Z, et al. Absence or mislocalization of DNAH5 is a characteristic marker for motile ciliary abnormality in nasal polyps. Laryngoscope. 2018;128(3):E97–E104. doi:10.1002/lary.26983
27. Al-Rawi MM, Edelstein DR, Erlandson RA. Changes in nasal epithelium in patients with severe chronic sinusitis: a clinicopathologic and electron microscopic study. Laryngoscope. 1998;108(12):1816–1823. doi:10.1097/00005537-199812000-00010
28. Hornef N, Olbrich H, Horvath J, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med. 2006;174(2):120–126. doi:10.1164/rccm.200601-084OC
29. Lee JH, Mcdonald ML, Cho MH, et al. DNAH5 is associated with total lung capacity in chronic obstructive pulmonary disease. Respir Res. 2014;15(1):97. doi:10.1186/s12931-014-0097-y
30. Yang Y, Song B, Yang X, et al. Predictive significance of enhanced level of angiogenesis and tissue neutrophils for antrochoanal polyps recurrence in children. Ear Nose Throat J. 2022;101(7):P284–P290. doi:10.1177/0145561320963627
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.