Back to Journals » International Journal of General Medicine » Volume 14
Amniocentesis and Next Generation Sequencing (NGS)-Based Noninvasive Prenatal DNA Testing (NIPT) for Prenatal Diagnosis of Fetal Chromosomal Disorders
Authors Qi QG, Tuo Y, Liu LX, Yu CX, Wu AN
Received 15 December 2020
Accepted for publication 15 April 2021
Published 11 May 2021 Volume 2021:14 Pages 1811—1817
DOI https://doi.org/10.2147/IJGM.S297585
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Qi-Ge Qi,1,* Ya Tuo,2,* Li-Xue Liu,2,* Cong-Xiang Yu,2,* Ai-Ning Wu2
1Department of Clinical Laboratory and Pathology, The 969th Hospital of P.L.A., Hohhot, Inner Mongolia, 010051, People’s Republic of China; 2Department of Obstetrics and Gynecology, The Affiliated Hospital of Inner Mongolia Medical University, Inner Mongolia University, Hohhot, Inner Mongolia, 010050, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ai-Ning Wu
Department of Obstetrics and Gynecology, The Affiliated Hospital of Inner Mongolia Medical University, Inner Mongolia University, No. 1 Of Tongdao North Road, Hui District, Hohhot, Inner Mongolia, 010050, People’s Republic of China
Tel +86 0471-6636630
Email [email protected]
Objective: The present study aimed to evaluate and analyze the results of karyotyping by amniocentesis and next generation sequencing (NGS)-based noninvasive prenatal DNA testing (NIPT) for the prenatal diagnosis of fetal chromosomal disorders.
Methods: A total of 2267 high-risk pregnant females with the indications for prenatal diagnosis who met the enrollment criteria between January 2015 and May 2019 at the Affiliated Hospital of Inner Mongolia Medical University were included and underwent NGS-based NIPT in the present study. Amniocentesis, chromosome karyotyping by cell culture, and follow-up of the pregnancy outcomes were also conducted in the NIPT-positive pregnant females to assess the consistency between NIPT and results of karyotyping by amniocentesis.
Results: Among the 2267 cases, 29 cases were positive for NIPT, including 10 cases with a high risk of trisomy 21, 2 cases with a high risk of trisomy 18, 2 cases with a high risk of chromosome 13, and 20 cases with sex chromosome abnormalities. All the above NIPT-positive cases underwent amniocentesis, and 20 cases were eventually diagnosed. The sensitivity and specificity of NIPT for the diagnosis of trisomy 21, trisomy 13, and trisomy 18 were 100%, 99.96%, 100%, and 99.96%, 100%, 100%, respectively, and the positive predictive values were 91.67%, 66.67%, and 100%, respectively.
Conclusion: NGS of the fetal free DNA from the peripheral blood of pregnant females was an important complement to the prenatal diagnosis of chromosomal disorders represented by fetal chromosome aneuploidy with high sensitivity and specificity. In combination with the traditional karyotyping by amniocentesis, it could improve the diagnostic efficacy for fetal chromosomal disorders.
Keywords: next generation sequencing, karyotyping by amniocentesis, prenatal diagnosis, prenatal screening, fetal free DNA
Introduction
China is a country with high incidence of birth defects, and data show1 that chromosomal abnormalities, as important genetic defects, may affect between 1/200 and 1/150 of the newborns. Knowing how to conduct appropriate prenatal screening and diagnosis of chromosomal disorders is important to reduce the incidence of birth defects. Currently, chromosome karyotyping by amniocentesis is the main method of prenatal diagnosis. However, as an invasive method, it inevitably increases the risk of intrauterine infection and miscarriage, resulting in low clinical acceptance. With the continuous improvement and comprehensive development of next generation sequencing (NGS), noninvasive prenatal DNA testing (NIPT) through the NGS of peripheral blood of pregnant females is becoming the new direction of prenatal screening for high-risk populations.2,3 Owing to the short development duration of the NGS technology in the field of prenatal diagnosis, the impact of NIPT on the traditional prenatal diagnosis needs to be further confirmed. The present study aimed to detect the fetal free DNA in the peripheral blood of 2267 high-risk pregnant females with indications for the prenatal diagnosis, and perform comparative analysis of the gold standard of karyotyping by amniocentesis to provide a reference for the application of a new model of prenatal screening for fetal chromosomal disorders.
Materials and Methods
General Material
A total of 2267 pregnant females who attended the Affiliated Hospital of Inner Mongolia Medical University between January 2015 and May 2019 were included in the present study. The age range was 22–47 years, with an average age of 31.7 ± 4.6 years. The gestational age was 12–26+6 weeks, and the number of pregnancies was 1–5. There were 1309 cases of primiparous females and 958 cases of postpartum females. The inclusion criteria were as follows: (1) females with natural conception; (2) those with the indications for prenatal diagnosis (including those with high risk and borderline risk in the serological screening for Down’s syndrome, those with ultrasonographic soft index abnormality, those with advanced age, and those with adverse birth history); (3) those who volunteered to participate in NIPT, and those for whom the diagnosis was provided by amniocentesis in the case of positivity; and (4) those who signed the informed consent form. The exclusion criteria were as follows: (1) those with a gestational age of <12+0 weeks; (2) those with BMI >40 kg/m2; (3) one of the couple had a clear chromosomal abnormality, and those with a recent history of allogeneic blood transfusion, immunotherapy, transplantation, or stem cell therapy; and (4) those with a history of exposure to other special teratogenic agents. The present study was approved by the Ethics Committee of the Affiliated Hospital of Inner Mongolia Medical University (Approval number: NO.CS2019003), and all the patients signed an informed consent form. This study was conducted in accordance with the Declaration of Helsinki.
Methods
Method of NIPT
The prenatal screening procedures were strictly implemented, and NIPT was conducted under the principle of genetic counseling and informed consent.
First, 5–10 mL of the peripheral blood was drawn from the pregnant females using the Ethylene Diamine Tetraacetic Acid blood collection tube. The samples were centrifuged to separate the plasma within 2 h (the parameters were as follows: speed 1600 r/min with a duration of 10 min the first time; speed 1600 r/min with a duration of 10 min the second time). The free fetal DNA was extracted from the maternal blood using the plasma free DNA extraction kit (BerryGenomics, Beijing, China) operated according to the manufacturer’s instructions. With successful extraction of the DNA, the samples were stored at −20°C for further detection (after DNA quantitative analysis, those with a concentration of 0.05–0.7 ng/μL and a volume of >35 μL were considered to be the qualified samples).
Fragment repair was conducted in the free DNA, with the addition of the base “A” to the 3ʹ end, and the tags and splices for sequencing and analysis were added by the ligase to constitute a standard library ready for sequencing. The concentration of the standard library was quantified by Polymerase Chain Reaction amplification and use of the standard curve. The standards to judge the standard curve were as follows: R2 >99%, slope >−3.6, efficiency >90%. With the purification of the library that had passed quality control, the samples were loaded on the NextSeqCN500 gene sequencer (BerryGenomics, Beijing, China) to undergo the sequencing. Each read in the sequencing was aligned with the human reference genome map to obtain the raw values for each chromosome, and the sequencing results were submitted for analysis.
Observation of chromosomal Z-values (calculated from the chromosome representative values and the formula for calculating the minimum GC content): the normal reference range for the chromosome trisomy 13 was −3.0–3.0; the normal reference range for the chromosome trisomy 18 was −3.0–3.0; and the normal reference range for the chromosome trisomy 21 was −3.0–3.0. Results greater than or equal to 3.0 or less than or equal to −3.0 indicated a positive result.3
Diagnosis of Karyotyping by Amniocentesis and Follow-Up
Amniocentesis and G band karyotyping after cell culture were recommended for pregnant females with positive results of fetal free DNA in the peripheral blood. Amniocentesis was conducted under the guidance of ultrasonography. First, 15–20 mL of amniotic fluid was extracted and placed in a centrifuge tube. After centrifugation at 2000 rpm for 7 min, the cell suspension was inoculated in three cultures, with the incubation and fluid exchange following the standard procedures. When more cells were growing and attaching to the wall and with the formation of 8–10 cell clones under the microscope, the cells were transferred into a centrifuge tube to prepare the cell suspension. With the G band demonstration, according to the criteria of the American College of Pathologists, the karyotyping should be counted in at least 20 nuclear fission phases with 5 karyotypes being analyzed, and the resolution in karyotyping should be at least 320 bands. The nuclear nomenclature was conducted according to the ISCN 2009 system. The results of karyotyping by amniocentesis were regarded as the gold standard. Induced labor was conducted in those with abnormal chromosomal karyotypes, while intensive follow-ups were performed in those with low risk until the completion of induced labor or the birth of the fetus.
Statistical Analysis
The Excel database (Microsoft, Redmond, DC, USA) was constructed. SPSS 19.0 software (SPSS Version 19.0, SPSS) was adopted for the data analysis. The countable data were expressed as cases or rates, and the χ2 test was used for comparison. P < 0.05 was considered statistically significant.
Results
Basic Information of the Participants
As shown in Table 1, there’s no significant difference between the NIPT+ group and NIPT- group in age, age of menarche, or number of pregnancies, but BMI is significantly higher in NIPT+ group than NIPT- group.
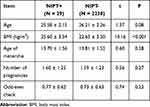 |
Table 1 Basic Information of the Participants |
Results of the Positivity in NIPT
Of the 2267 cases enrolled in the present study, 1185 cases had serologically high risk or borderline risk, accounting for 52.27%. In addition, there were 635 cases with advanced age, accounting for 28.01%. Furthermore, 386 cases had ultrasonographic abnormalities, accounting for 17.03%. Moreover, 61 cases had adverse birth histories, accounting for 2.69%. Using the NIPT screening, 29 positive specimens were detected in the 2267 specimens, with a positive detection rate of 1.28% (29/2267). Among them, 3 cases were trisomy 13, 12 cases were trisomy 21, 2 cases were trisomy 18, 4 cases were 45, XO, 6 cases were 47, XXX/XXY, and 2 cases were 47, XYY. The NIPT positive results and detection rates for different groups by the high-risk screening factors are shown in Table 2.
 |
Table 2 Results of Prenatal NIPT (Case) |
Comparison of Karyotyping by Amniocentesis and NIPT
All 29 pregnant females with positive NIPT results voluntarily underwent amniocentesis, and chromosome karyotyping by cell culture revealed 23 cases of chromosomal abnormalities. Among these cases, 2 cases of trisomy 18 and 2 cases of 47, XYY detected by NIPT were consistent with the results of cellular karyotyping. Of the 12 cases of trisomy 21, one case was revealed to be normal by karyotyping. Of the 3 cases of trisomy 13, one case was revealed to be 47, XXY by karyotyping. Of the 4 cases of 45, XO, the results of karyotyping revealed three cases to be normal and one case to be 47, XXX. The results of karyotyping in 6 cases of 47, XXX/XXY revealed one case to be normal. In the present study, induced labor was conducted in those with results of chromosome disorders in both the amniocentesis and NIPT. The details are provided in Table 3.
 |
Table 3 Comparison of the NIPT Results and Karyotyping by Amniocentesis |
Performance Analysis of NIPT for Prenatal Diagnosis
In the present study, the diagnostic performance of NIPT for trisomy 21, trisomy 13, and trisomy 18 in 2267 samples was analyzed. The results indicated that the sensitivity and specificity of NIPT for the diagnosis of trisomy 21, trisomy 13, and trisomy 18 were 100%, 99.96%, and 100%, and 99.96%, 100%, and 100%, respectively, with positive predictive values of 91.67%, 66.67%, and 100%, respectively. The results are presented in Table 4.
 |
Table 4 Results of NIPT in 2267 Cases |
Discussion
According to the traditional cytogenetic testing and analysis, the chromosome number abnormalities in genetic mutations include the polyploid and aneuploid. Chromosomal aneuploidy is the most common chromosomal disease, accounting for 80–90% of all chromosomal diseases,4 and it is the main cause of birth defects. Different types of fetal chromosomal aneuploidy have different prognoses. Poor prognosis, organ abnormalities, mental retardation, and growth retardation are the common features.3–5 Prenatal diagnosis is based on providing genetic counseling for high-risk pregnant females, with further applications of modern biology, biochemistry, immunogenetics, cytogenetics, and molecular genetics techniques to perform maternal or embryo/fetal testing to achieve the diagnosis of chromosomal abnormalities. Traditionally, among the procedures of prenatal diagnosis, the most commonly used and effective method of prenatal diagnosis for fetuses in mid-term pregnancy is amniocentesis and karyotyping of amniotic fluid cells. The genetic examination of the amniotic fluid cells can detect the whole chromosome abnormalities or a wide range of chromosomal variations with good diagnostic accuracy, which is conducive to the timely detection of fetal chromosomal abnormalities. However, there are strict indications and contraindications for amniocentesis. Pregnant females with placenta previa, threatened abortion, or active hepatitis are not suitable for the prenatal diagnosis by the amniotic fluid.6,7 This method may bring the risk of intrauterine infection of the fetus and leakage of the amniotic fluid, together with an increase in the psychological pressure of pregnant females, and therefore the acceptance rate is not high among high-risk pregnant females. Other traditional screening procedures, such as chorionic biopsy and cord blood puncture, are also invasive and have detection limitations, which limits the clinical application to a certain extent. Therefore, seeking a reasonable and practical prenatal screening and diagnostic method to achieve accurate detection of fetal chromosomal diseases is of great significance for reducing the incidence of birth defects.
With the development of modern molecular biology, the high-throughput sequencing technology (also known as NGS) is profoundly affecting the development status of genetics. Currently, NGS is being applied in the field of clinical genetic testing. The NGS represented by NIPT, through the noninvasive method, amplifying and counting of the free fetal DNA fragments in the maternal plasma, is used for detection of the genetic material nucleotides to realize the bioinformatics interpretation of the DNA fragments.2,3,6 NIPT is currently regarded as a breakthrough method for the detection of fetal chromosomal aneuploidy. A series of clinical verifications have confirmed that NIPT has high sensitivity and specificity for trisomy 21 and trisomy 18. Some scholars believe that the detection rate of NIPT in trisomy 21 is greater 98%, and the detection rate for trisomy 18 and trisomy 13 is greater than 90%.8,9 A retrospective study conducted by Greene et al10 confirmed that the implementation of NIPT considerably reduced the number of high-risk pregnant females who gave up further prenatal diagnosis and substantially improved the efficiency of prenatal diagnosis of fetal chromosomal aneuploidy. Vora et al11 reported that compared with those of karyotyping, the specificity and sensitivity of NIPT had higher consistency with the results of chromosome karyotyping to make better judgments on the fetal chromosomes. A study conducted by Lori et al12 on pregnant females with a high risk of chromosomal disorders in the second trimester showed that the sensitivity of NIPT for trisomy 21, trisomy 18, and trisomy 13 was 99.8%, 97.7%, and 97.5%, respectively. The specificity for trisomy 21, trisomy 18, and trisomy 13 was greater than 99.0%. There are also an increasing number of domestic studies2,6,13,14 showing that NIPT based on NGS has the advantages of noninvasiveness, high sensitivity for the diagnosis of aneuploidy, and good reproducibility of results. It can make a judgment of the chromosomal disorders earlier than invasive diagnosis such as amniocentesis. Although there is some degree of false positivity, the false positivity is very low (0.1–0.5%). NIPT based on NGS is especially suitable for the population with contraindications for invasive prenatal diagnosis or pregnant females aged over 35 years.
The results of the present study revealed that 29 cases (0.98%) were positive for the chromosomal diseases detected by NIPT with a positive detection rate of 1.28%, including 3 cases of trisomy 13, 12 cases of trisomy 21, 2 cases of trisomy 18, 4 cases of 45, XO, 6 cases of 47, XXX/XXY, and 2 cases of 47, XYY. The above results were consistent with the results of chromosome karyotyping of amniotic fluid. By comparing the above results with the results of chromosome karyotyping of amniotic fluid, the sensitivity and specificity of NIPT in the present study were 100%, 99.96%, and 100%, and 99.96%, 100%, and 100% for trisomy 21, trisomy 13, and trisomy 18, respectively, and the positive predictive values were 91.67%, 66.67%, and 100% for trisomy 21, trisomy 13, and trisomy 18, respectively. In the present study, the false positivity of NIPT for trisomy 21 was 0.04%, and there was one case with the results of NIPT suggesting trisomy 21, which was determined to be normal by karyotyping. In another case, the result of NIPT was trisomy 13 while the karyotyping result was 47, XXY, indicating that for chromosomal abnormalities, there was still a certain degree of false positivity in NIPT, which was consistent with the findings reported in the literature. A study15 has shown that the cffDNA was derived from the placental trophoblasts and that the sequencing depth during the NGS might lead to sequencing bias and subsequent pairwise splicing, which, in turn, would affect the statistical analysis. However, the factors influencing the false positivity results of NIPT tests need further investigation. In the present study, there were four cases of chromosomal abnormalities with a genetic test of 45, XO, while the karyotyping of the amniotic fluid cells confirmed three cases of normal karyotype and one case of 47, XXX. The results of NIPT showed six cases of 47, XXX/XXY, while after karyotyping, one case was considered to be normal. These indicated that there was slightly higher false positivity of NIPT for the diagnosis of sex chromosome aneuploidy. The results of the present study indicated that NGS might be suboptimal in identifying the risk of sex chromosome aneuploidy abnormalities, which might be related to the small sample size and the methodological limitations of NIPT. Generally, it is believed that the diversity of karyotypic abnormalities and the complexity of clinical manifestations of sex chromosomes are the main reasons for the false positivity or uncertain results of the high-risk sex chromosomes by NIPT. Moreover, placental restriction of chimerism, fetal chimerism, and maternal chimerism can affect the results of fetal sex chromosome analysis. Therefore, some clinical experts still have doubts about the use of NIPT in the detection of sex chromosome abnormalities, abnormal chromosomal balance structures (translocation, retroversion), and chimeric chromosomes. Some experts16,17 have argued that the specificity and accuracy of NIPT for sex chromosomes and other chromosomes is low and that NIPT should mainly be used as a screening method for aneuploidy of the chromosome of trisomy 21, trisomy 18, and trisomy 13. The selection of indications for NIPT for prenatal diagnosis of chromosomal disorders needs to be further explored. However, we did not detect fetal fractions and exact sequencing depth, nor the distributions of Z-scores of both negative and positive samples, which are important for both sensitivity and specificity of the Z-score method.
In summary, it was further demonstrated by the present study that NGS of the fetal free DNA from the peripheral blood of the pregnant females could be used for the prenatal diagnosis of fetal chromosomal disorders in high-risk pregnant females with high sensitivity and specificity. Moreover, it might be a good screening method for chromosomal aneuploidy, which could be an important supplement to the prenatal diagnosis. However, there was a certain degree of false positivity in NIPT, and when the NIPT result was abnormal, it still needed to be combined with traditional karyotyping of the amniotic fluid cells to improve the diagnostic performance on fetal chromosomal disorders.
Acknowledgments
We would like to acknowledge the hard and dedicated work of all the staff that implemented the intervention and evaluation components of the study.
Funding
National Natural Science Foundation of China (81660542); Science and Technology Planning Project of Inner Mongolia Autonomous Region, (2019GG155).
Disclosure
The authors declare that they have no competing interests.
References
1. Vong JSL, Jiang P, Cheng SH, et al. Enrichment of fetal and maternal long cell-free DNA fragments from maternal plasma following DNA repair. Prenat Diagn. 2019;39(2):88–99. doi:10.1002/pd.5406
2. Zhu L, Cheng J, Zhou B, et al. Diagnosis for choroideremia in a large Chinese pedigree by next-generation sequencing (NGS) and non-invasive prenatal testing (NIPT). Mol Med Rep. 2017;15(3):1157–1164. doi:10.3892/mmr.2017.6119
3. Keravnou A, Ioannides M, Loizides C, et al. MeDIP combined with in-solution targeted enrichment followed by NGS: inter-individual methylation variability of fetal-specific biomarkers and their implementation in a proof of concept study for NIPT. PLoS One. 2018;13(6):1–13. doi:10.1371/journal.pone.0199010
4. Koumbaris G, Achilleos A, Nicolaou M, et al. Targeted capture enrichment followed by NGS: development and validation of a single comprehensive NIPT for chromosomal aneuploidies, microdeletion syndromes and monogenic diseases(Article). Mol Cytogenet. 2019;12(1). doi:10.1186/s13039-019-0459-8
5. Feng X, Shen H, Ge Y, et al. Prenatal screening and prenatal diagnosis of down syndrome and laboratory quality control. Chin J Eugen Genet. 2019;27(08):944–945.
6. Lin YY, Xu WF, Liu YH, et al. Application of noninvasive prenatal testing based on massively parallel sequencing for the detection of chromosomal abnormality. Chin J Birth Health Heredity. 2016;24(01):52–54.
7. Yang XK, Guo XL, Zhong J, et al. Noninvasive prenatal genetic testing in 6804 pregnant women aged less than 35 years with positive results in serum screening. J South Med Univ. 2019;39(11):1350–1356. doi:10.12122/j.issn.1673-4254.2019.11.13
8. Canick JA, Lambert M, Lian GM, et al. Advances in ultrasound combined with noninvasive prenatal DNA testing in the screening of fetal chromosomal abnormalities in early pregnancy. Mol Cytogenet. 2018;31(12):1814–1815.
9. Yannis D, Claire B, Valérie M. Quality control of prenatal screening and diagnosis for cytogenetic testing. Mol Genet Genom Med. 2020;8(3):251–256.
10. Greene MF, Phimistereg E. Screening for trisomies in cir-culating DNA. N Engl Med. 2014;370(9):874–875. doi:10.1056/NEJMe1401129
11. Vora NL, Robinson S, Hardisty EE, Stamilio DM. Utility of ultrasound examination at 10–14 weeks prior to cell-free DNA screening for fetal aneuploidy. Ultrasound Obstetr Gynecol. 2017;49(4):465–469. doi:10.1002/uog.15995
12. Dobson LJ, Reiff ES, Little SE, Wilkins-Haug L, Bromley B. Patient choice and clinical outcomes following positive noninvasive prenatal screening for aneuploidy with cell‐free DNA (cfDNA). Prenat Diagn. 2016;36(5):456–462. doi:10.1002/pd.4805
13. Zheng YY, Wan XN, Song TT, et al. Clinical application of noninvasive DNA prenatal testing in screening fetal chromosome Aneuploidy in 8594 pregnant women. J Pract Obstetr Gynecol. 2019;35(01):68–71.
14. Fu J, Guo HC, Xiao JP, et al. The clinical application of noninvasive prenatal DNA screening in fetal chromosome aneuploidy gene detection.Chinese. J Fam Plan. 2018;26(11):1064–1071.
15. Miltoft CB, Rode L, Bundgaard JR, Johansen P, Tabor A. Cell-free fetal DNA in the early and late first trimester. Fetal Diagn Ther. 2020;47(3):1–9. doi:10.1159/000502179
16. Palomaki GE, Kloza EM. Prenatal cell-free DNA screening test failures: a systematic review of failure rates, risks of down syndrome, and impact of repeat testing. Genet Med. 2018;20(11):1312–1323. doi:10.1038/gim.2018.22
17. Lambert-Messerlian GM, Eklund EE, Neveux LM, Palomaki GE. Measuring maternal serum screening markers for Down’s syndrome in plasma collected for cell-free DNA testing(Article). J Med Screen. 2017;24(3):113–119. doi:10.1177/0969141316670193
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
