Back to Journals » The Application of Clinical Genetics » Volume 8
Alström syndrome: current perspectives
Authors Álvarez-Satta M, Castro-Sánchez S, Valverde D
Received 23 April 2015
Accepted for publication 22 May 2015
Published 21 July 2015 Volume 2015:8 Pages 171—179
DOI https://doi.org/10.2147/TACG.S56612
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Martin Maurer
María Álvarez-Satta, Sheila Castro-Sánchez, Diana Valverde
Departamento de Bioquímica, Genética e Inmunología, Facultad de Biología, Universidad de Vigo, Vigo, Spain
Abstract: Alström syndrome (ALMS) is a rare genetic disorder that has been included in the ciliopathies group, in the last few years. Ciliopathies are a growing group of diseases associated with defects in ciliary structure and function. The development of more powerful genetic approaches has been replaced the strategies to follow for getting a successful molecular diagnosis for these patients, especially for those without the typical ALMS phenotype. In an effort to deepen the understanding of the pathogenesis of ALMS disease, much work has been done, in order to establish the biological implication of ALMS1 protein, which is still being elucidated. In addition to its role in ciliary function and structure maintenance, this protein has been implicated in intracellular trafficking, regulation of cilia signaling pathways, and cellular differentiation, among others. All these progresses will lead to identifying therapeutic targets, thus opening the way to future personalized therapies for human ciliopathies.
Keywords: Alström syndrome, ciliopathies, ALMS1 gene, ALMS1 protein, molecular diagnosis
Clinical findings
Alström syndrome (ALMS, OMIM 203800) is a very rare autosomal recessive disease. Alström et al described the clinical features of three patients in a paper published in 1959,1 in which they give a detailed phenotype of these patients, showing a combination, inherited and recessive, of retinal degeneration, obesity, neurosensorial deafness, and type 2 diabetes mellitus (T2DM). The estimated prevalence for ALMS is one to nine cases per 1,000,000 individuals2 with nearly 700 cases described worldwide to date. Until now, disease-causing mutations in the ALMS1 gene have been involved in this disorder.
First symptoms appear in childhood with a great variability in terms of severity and clinical evolution. This variability has been described among patients with identical mutations within the same family.3,4 The severity of the symptoms of this syndrome often result in organ failure, drastically reducing life expectancy which rarely exceeds 50 years. Table 1 summarizes the most relevant clinical features related to ALMS.
  | Table 1 Summary of clinical findings frequently observed in Alström syndrome patients |
Photoreceptor dystrophy, present in 100% of patients with ALMS, starts to develop between birth and the first 15 months of life. It is normally accompanied by photophobia and nystagmus.5 An initial loss of cone function followed by a loss in rod function occurs, leading to an early blindness.6 Another important feature of ALMS is progressive sensorineural hearing loss, which occurs in the first decade of life in 70% of patients. This defect may progress to a moderately severe hearing loss or deafness, between the first and second decades of life.7 All these changes in the neurosensory capabilities at an early age have important consequences not only on the social development of children, but also in their adaptation to the external environment.
In the first 5 years of life, many ALMS patients develop truncal obesity, insulin resistance, hyperinsulinemia, and hyperlipidemia.8 These symptoms may progress to T2DM usually accompanied by acanthosis nigricans (a rare skin disorder characterized by the presence of hyperkeratosis and hyperpigmentation in skin folds and flexible body areas). Interestingly, despite the early development of T2DM, patients with ALMS do not show the typical diabetic peripheral neuropathy. This fact suggests that mutations in ALMS1 could somehow, still unexplained, protect the development of neuropathy induced by hyperglycemia of T2DM.9 Hypertriglyceridemia is another common clinical feature in this syndrome and can lead to acute pancreatitis.10,11
ALMS patients may also show infertility caused by hypogonadism (especially in men). Women may have polycystic ovarian syndrome, hirsutism, and insulin-resistant hyperandrogenism.12 Other endocrine disorders observed in ALMS are hypothyroidism, changes in the age of onset of puberty, and short stature associated with alterations in the growth hormone/insulin-like growth factor 1 axis.7,13–15 Dilated cardiomyopathy or congestive heart failure occurs in approximately 70% of patients during childhood or adolescence, being a frequent cause of death.5,16,17
Other common features in ALMS are renal and pulmonary failure, and hepatic dysfunction.7 Autopsies of patients often exhibit multiple organ fibrosis in liver, kidney, ovaries, and seminiferous tubules.7 The origin of this fibrosis and its contribution to multiorgan failure of this syndrome is unknown, although some progress has been recently made.18
Although cognitive impairment is not a common feature of ALMS, approximately 45% of affected children have some delayed learning skills or milestones.19 Absence seizures and general sleep disturbances may be included among other neurological manifestations of ALMS.7 The frequency of psychiatric disorders in individuals affected by ALMS has not been determined.
Marshall et al reported an in-depth revision of the ALMS clinical phenotype.5 However, some atypical, extreme cases with mutations in the ALMS1 gene have been described ever since, either without nystagmus, photophobia, obesity, and hearing loss or only with mitogenic cardiomyopathy.20–22 Therefore, the clinical definition of the syndrome needs to be revised and extended.
On the other hand, Ozantürk et al reported clinical and molecular findings in a large Turkish cohort of ALMS patients.23 Although the clinical spectrum did not differ from other clinical descriptions of the syndrome, they described some cases of triallelism associated with ALMS (three pathogenic mutations in the same gene). This triallelic condition did not involve a more severe phenotype, but functional analyses of these mutations will be needed, in order to elucidate their role in ALMS1 protein function.
Early diagnosis of ALMS is complicated by the progressive onset of the associated symptoms, as well as its own inter- and intrafamilial clinical heterogeneity, as frequently occurs in other ciliopathies such as Bardet–Biedl syndrome (BBS). It is set based on the phenotype of the patient, and the diagnosis is confirmed when the molecular analysis identifies two mutations in ALMS1 gene.5
Molecular genetics
ALMS1 gene is located on chromosome 2p13 and consists of 23 exons that are predicted to encode a large protein of 4,169 amino acids,24 whose biological role is still being elucidated (Figure 1). To date, several mutations have been described in this gene, being exons 8, 10, and 16 hotspots for ALMS1 mutations (variations in exon 8 account for 49% of the total mutation load in ALMS).25 Nevertheless, slightly different percentages have been reported in Turkish patients, being mutations in exons 8 and 10 detected in 72% of the cases. It has to be taken into account that most of the affected individuals were homozygous, or compound heterozygous, but some cases of triallelism have also been found.23 More than 200 mutations have been reported, the vast majority involving nonsense or frameshift mutations that lead to a predicted premature stop codon.25 It has been proposed that ALMS could be underdiagnosed, as some mutations that predict a premature termination appear in the general population and might not be disease-causing alleles.5 To date, atypical phenotypes have been described as well, supporting the hypothesis that ALMS phenotypes are strongly modified by the genetic background and could result in different clinical phenotypes.
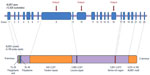  | Figure 1 ALMS1 gene (top) and ALMS1 protein. (bottom). |
ALMS belongs to a growing class of human diseases with overlapping phenotypes, referred as ciliopathies. These include disorders such as BBS and Senior–Løken syndrome. The clinical similarity among these syndromes, particularly with BBS, and the delayed onset of some clinical features in ALMS sometimes result in misdiagnosis.26
New genetic approaches in molecular diagnosis
In recent years, the number of patients with a confirmed molecular diagnosis has been growing mainly due to the advent of more powerful genetic tools, especially next-generation sequencing (NGS) technology. These genetic approaches represent a very important breakthrough, above all for those patients who do not show the typical ALMS phenotype, since they could suffer from long-term complications associated with a delayed diagnosis of this pathology.27
The first strategy followed in most of the cases is to discard the presence of mutations in ALMS1 gene hotspots (exons 8, 10, and 16), since a complete sequencing of this gene is very tedious due to its size. Although the arrayed primer extension technology (which includes a set of known mutations in ALMS1 and ten BBS genes) was proposed as a relatively cost-effective first approach for molecular diagnosis,28 the growing development of NGS is rapidly replacing the previous approaches.23 One of these new methods consists of specific panels designed for testing a set of genes of interest,5 but currently the most widely used molecular approaches are whole-exome sequencing (WES), and whole-genome sequencing. This is due mainly to the reduction of their costs, enabling an in-depth analysis in a considerable number of patients at affordable prices, and also discards mutations in other genes.27,29
As mentioned earlier, some studies have suggested that many of nonclassic ALMS cases could go unnoticed, taking into account the current diagnostic criteria and if the wrong molecular strategy is followed.20 It has also been proposed that a different location or nature of the ALMS1 mutations may influence the presence of nonclassic clinical features in these patients.20 An additional problem to be considered is the overlapping phenotypes among different ciliopathies, particularly with BBS in the case of ALMS, which further complicates a proper molecular diagnosis.5
The main advantage of WES approach lies in its broad analysis of all coding regions in the genome, detecting the disease-causing mutations in a rapid and easier way.29 NGS technology also provides the possibility of identifying novel genes involved in this group of pathologies. Though the interpretation of exome/genome data is a challenging task due to the number of regions analyzed (size), this problem is being partially solved by the enhanced bioinformatics tools which facilitate the variant filtering, a critical point for identifying the disease-causing gene.30
WES has been able to elucidate some intriguing phenotypes, which in principle did not seem to be associated with the typical ALMS phenotype but finally resulted in mutations in ALMS1 gene. Thus, WES has solved the limitation of genetic panels in a sporadic patient suffering from familial dilated cardiomyopathy and severe heart failure, identifying pathogenic mutations in compound heterozygous state in ALMS1 gene.30 In addition to this, it has been reported that the combination of WES and a previous linkage analysis has facilitated the identification of pathogenic mutations in ALMS1 gene in a consanguineous Turkish family with severe dilated cardiomyopathy as well.22 This family was not previously suspected to be an ALMS case because of the absence of several features typically related to this syndrome.22 On the other hand, homozygosity mapping followed by WES was another strategy carried out in consanguineous Leber congenital amaurosis families, unexpectedly resulting in the identification of ALMS1 mutations in these families.31 These studies underline the perspectives offered by the combination of different genetic tools as a powerful approach, especially in the case of consanguineous families, or those who present nontypical ALMS phenotype, underscoring again the genetic and clinical overlapping among ciliopathies.
Taking into account the aforementioned, we could say that the combination of an accurate molecular diagnosis and a complete clinical phenotype represents an indispensable step to devise a future appropriate treatment and management of the pathology, in which NGS technologies will be critical for the ALMS diagnosis as cost-effective tools when compared with traditional polymerase chain reaction and direct Sanger sequencing.31
ALMS as a ciliopathy: therapy options
Clinical manifestations associated with the different ciliopathies can be present in almost all tissue throughout development, including sensorial dysfunction as well as defects in multiple organs.32 Despite new ciliary genes being discovered through the identification of new disease-causative mutations in genes coding for ciliary proteins, and also delving deeper into the molecular basis of these pathologies, the current measures of therapy for these patients are limited, without real curative options. Hence, it is important to search for an effective treatment for these patients in order to reduce or limit the disease progression.32
In recent years, several pharmacological trials have been carried out in some ciliopathies, mainly focused on recovering renal or hepatic function, as in the case of polycystic kidney disease (PKD),33,34 as well as on delaying retinal degeneration progression in BBS,35 so its efficacy in other affected organs is limited. Therefore (and given that up to now the main options for the majority of patients are restricted to the management of the clinical symptoms and the improvement of the quality of life), it remains essential to continue to make progress in the knowledge of the pathophysiological basis of these diseases, characterizing the role of the proteins involved so that it allows us to identify therapeutic targets.32 To this end, the ongoing identification of novel ciliary genes, mainly due to NGS technology, represents the starting point for the development of individualized gene therapies for these ciliopathies patients.
Biological roles of ALMS1 protein: current knowledge
The study of ALMS1 protein has focused the interest of many researchers since ALMS gives a monogenic model of obesity and metabolic syndrome.5 ALMS1 is a protein of 4,169 amino acids that localizes in centrosomes and basal bodies of primary cilia.36–38 The biological functions of this protein are still being elucidated, and encompass ciliary function and structure maintenance, intracellular trafficking, regulation of cilia signaling pathways, and cellular differentiation, among others (Figure 2).
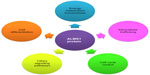  | Figure 2 Schematic view of biological functions presumably related to ALMS1 protein until now. |
Looking into the biological role of ALMS1 protein is already providing us with challenging findings not only regarding cilia biology but also the involvement of primary cilia in cellular processes that had not been previously linked to ciliary proteins.
The role of ALMS1 protein in energy/metabolic homeostasis
Over the last few years, pivotal progress has been achieved in unraveling the ALMS1 involvement in energy balance and appetite regulation, whose altered regulation leads to the development of obesity and diabetes, both features classically associated to ALMS patients.
In this sense, Heydet et al reported a role of ALMS1 in satiety regulation in the obese mouse strain foz/foz, harboring a truncating mutation in ALMS1 gene.39 Thus, the basal body localization of this protein in hypothalamic neurons of control mice is not found in model animals. This finding, altogether with the strong reduction of hypothalamic neurons at postnatal period in model mice and the significant loss of primary cilia involved in appetite regulation, suggests a crucial role of ALMS1 in the maintenance and stability of cilia structure and/or function in these neurons.39
This study also points out that different molecular etiologies for satiety alterations in BBS compared with ALMS may exist. Hence, the localization of two well-known proteins involved in hypothalamic appetite regulation (melanin-concentrating hormone receptor 1 and somatostatin receptor type 3) is conserved in foz/foz animals but lost in Bbs2 and Bbs4 knockout mice models, suggesting different roles for BBS and ALMS1 proteins.39
However, clearer evidence of metabolic dysfunction caused by ALMS1 deficiency was recently reported.40 The authors describe an altered glucose transporter 4 (GLUT4) trafficking in the Alms1GT/GT mouse model, which suggests a role of ALMS1 in glucose homeostasis through GLUT4 trafficking pathway. Thus, early signs of metabolic dysfunction such as reduced total GLUT4 content and altered translocation to the plasma membrane were observed in model mice. It is noteworthy to highlight that the observation of strong glucose intolerance prior to metabolic disease in Alms1GT/GT mice suggests the existence of one or several underlying, not yet defined compensatory mechanisms, maybe in brown adipose tissue or muscle, which will have to be elucidated.40
ALMS1 protein in intracellular trafficking and cell cycle control
Many efforts focused on elucidating the biological role of ALMS1 related to its involvement in cell cycle control and intracellular trafficking regulation. One of the first studies which made remarkable findings was published by Zulato et al, with special relevance in the context of the widespread fibrosis, frequently developed by ALMS patients, which is often related to a premature death.18 This work referred to significant differences in expression levels for more than 500 genes belonging to functional categories such as cell cycle, apoptosis, cellular motility, and extracellular matrix, after performing gene expression analysis in dermal fibroblasts from ALMS patients. Genes coding for extracellular matrix components and their regulators (especially, those related to collagen expression and secretion) were observed to be strongly upregulated. In addition, cultured fibroblasts from ALMS patients showed alterations in cytoskeleton and morphology, and also displayed a longer cell cycle and an increased resistance to apoptosis. Altogether, these results would support the emerging role of ALMS1 as key regulator in cell and extracellular matrix normal activity and interactions.18
To gain better understanding of ALMS1 biological role, the implication of this protein in the recycling endosome pathway was studied.41 They found several interacting partners with the ALMS1 C-terminus, mainly α-actinins and other proteins previously linked to endosome trafficking. Moreover, patient’s fibroblasts showed a lower endocytic activity and an altered morphology of actin fibers, which suggest that aberrant protein recycling processes may underlie ALMS pathogenesis. With this in mind, it is feasible to think that any disruption in the cytoskeleton architecture organization, which is required to proper endosomal trafficking, could exist in ALMS patients as proteins belonging to cytoskeleton-associated recycling or transport complex were identified as ALMS1-interacting proteins.41 Furthermore, this complex is required for the receptor recycling from early endosomes to the plasma membrane, what would support this hypothesis.41 This body of evidence is opening new stimulating research lines which will require further investigation to enlarge an accurate knowledge regarding ALMS1 biology.
It is worth highlighting the growing evidence that link endocytic recycling deficiency to metabolic alterations commonly observed as part of ALMS phenotype, such as impaired glucose uptake. For example, α-actinin-4 directly interacts with ALMS1 and has been associated to GLUT4 trafficking which suggests a probably role of ALMS1 in the movement of GLUT4 vesicles along actin filaments to the plasma membrane.40–42
Finally, it is also important to note that the probable role of ALMS1 in cytokinesis is directly related to the endosome recycling pathway.41 Thus, this protein could be involved in direct trafficking of membrane components to the cleavage furrow of dividing cells.
Role of ALMS1 protein in signaling pathways coordinated by the primary cilium
The role of primary cilium as a coordinator of a wide variety of signaling pathways involved in cell cycle control, differentiation, and other cellular processes during development and tissue homeostasis, has been extensively studied for a number of years.43–46 However, ALMS1 involvement has not yet been thoroughly investigated, unlike BBS proteins which are well known to be linked to Sonic hedgehog and Wnt (Wingless+int-1) signal transduction pathways, among others.46–48
In this sense, evidence of ALMS1 as regulator of ciliary signaling networks is still being gathering. The role of this protein in cochlea development has been explored, as progressive sensorineural hearing loss is one of the ALMS distinctive clinical features.49 The basal body localization of ALMS1 in hair cells, the retained centriolar ALMS1 expression into maturity, and the cochlear histopathology observed in model mice pointed out a role in cilium-dependent planar cell polarity signaling. It would be specifically related to the basal body migration and/or anchoring during determination of final planar polarization of the cell.49
Notwithstanding, the strongest support until now was given by Leitch et al, who referred the regulation of Notch signaling via proper endosomal trafficking by basal body proteins such as BBS1, BBS4, and ALMS1.50 Hence, loss of these proteins led to pathway overactivation and receptor accumulation in late endosomes, providing a new link between basal body proteins and endosomal recycling, as already suggested.41 Remarkably, this study50 describes different trafficking defects caused by BBS/ALMS1 proteins, since loss of BBS proteins resulted in impaired Notch receptor recycling, whereas ALMS1 loss altered neither localization nor recycling of the receptor. Misregulation of this signaling pathway may provide a new mechanism that explains the development of BBS/ALMS phenotypes, given that Notch is known to be involved in neurogenesis and renal defects, frequently observed in the clinical spectrum of ciliopathies.50
Involvement of ALMS1 protein in cell differentiation
As in the previous case, ALMS1 role in cellular differentiation has not been deeply examined, but several recent studies report an emerging role of this protein at least in some cell types. For example, ALMS1-mediated adipocyte differentiation was suggested, since an early decrease in Alms1 mRNA expression during preadipocyte to mature adipocyte conversion in mouse 3T3-L1 cells has been detected, although it remained unaltered when fat cell insulin sensitivity changed.51 On the other hand, adipogenesis and insulin-stimulated glucose uptake were observed to be severely impaired when Alms1 expression was knocked down in adipocytes from 3T3-L1 cells.52 All these findings provide evidence of an altered adipocyte differentiation as possible cause, at least partially, of metabolic phenotype displayed by ALMS patients, likewise previously established for BBS.53
However, cultured preadipocytes isolated from Alms1GT/GT mouse model, which recapitulates the human ALMS phenotype, did not display abnormal adipogenic differentiation but mature adipocytes showed a reduced insulin-stimulated glucose uptake.40 Interestingly, Alms1 expression was abundant only in control cells, whereas GLUT4 mRNA levels were significantly lower in mature adipocytes from Alms1GT/GT animals. This suggests that ALMS1 is not directly involved in proper adipocyte maturation in vivo, so that intricate interactions may be required to expansion of adipose tissue in animals.40 These observations are in contrast with those given by Huang-Doran and Semple, which cannot explain the obese phenotype observed in ALMS models and patients.40,52
More recently, mutations in ALMS1 have been involved in cardiomyogenesis after performing WES in several children suffering from mitogenic cardiomyopathy.21,22 When inhibiting Alms1 mRNA expression in cultured neonatal murine cardiomyocytes, higher levels of cardiomyocyte proliferation markers were observed, suggesting a role of ALMS1 in cardiomyocyte cell cycle arrest during postnatal period.21 Thus, this protein could be considered as a key molecule for cell cycle regulation in perinatal cardiomyocites, activating the Wnt/β-catenin signaling pathway, known to be involved in cardiomyogenesis, when ALMS1 levels decreased.21 In this regard, another study describes that periostin overexpression found in cultured dermal fibroblasts from ALMS patients might be pointing out some involvement of this extracellular matrix protein in cardiac remodeling in ALMS.18
Conclusion and perspectives
The great progress made to unravel the pathogenesis of ALMS by different multidisciplinary groups in the last years is giving rise to promising advances in knowledge of its molecular basis. Nevertheless, the complex pathology of this kind of syndromes, with several organs affected, implies that the underlying molecular mechanisms remain to be clarified.
All this information will lead to establish molecular hotspots that could represent therapeutic targets in the future, the ultimate goal of all the research on rare diseases. However, it should not be forgotten, that the phenotypic complexity of ALMS, like most of ciliopathies, makes the real option of gene therapy very difficult, at least in adults. Hence, it is extremely necessary to continue exploring the underlying pathogenesis of ALMS disease. Maybe alternative mechanisms such as altered noncoding RNA expression patterns could be also contributing to ALMS as recently suggested,54 which may suppose an easier approach to gene therapy in these patients. Regarding this, DNA methylation can not be excluded as contributor to ALMS disease, since it has been involved in autosomal dominant PKD, another ciliary disorder.55
Several studies have demonstrated the success of some gene therapy techniques in restoring cilia dysfunction, all of them based on introducing wild-type genes via viral delivery. In the case of PKD, the restoration of cilia motility has been possible via lentiviral transduction of wild-type DNAI1 gene in respiratory epithelial cells.56 In addition, the use of adenoviruses has allowed to restore Ift88 gene expression, producing functional olfactory cilia which were absent due to a mutation responsible for the development of Oak Ridge Polycystic Kidney disease, what could be applied to other human ciliopathies.57 Protein trafficking can also be ablated in some ciliopathies, leading to defects in ciliary signaling.58 According to this, it has been shown that BBS mutations can produce structural changes in the corresponding protein, leading to an aberrant subcellular localization.32 This problem has also been solved in a BBS4 mutated gene mice, improving their retinal responses due to the restoration of rhodopsin localization.59
However, all the previous techniques have some limitations, such as gene size, overexpression, or cell type specificity, which can be solved with a direct manipulation of the endogenous mutated gene, either by repairing DNA mutations or manipulating the splicing mechanism.32 Furthermore, despite the importance of pharmacological treatments, the hope of real curative options lies in gene therapy or prospective treatments with stem cells, given its potential to offer individual options by means of targeting specific tissues or organs with long-lasting effectiveness.32
Taking all this in mind, we could say that there is still a long way to go to develop effective gene therapy treatments for human ciliopathies, but these first steps establish the starting point to achieve it. Moreover, exciting perspectives are opening up to determine the biological role of ALMS1, but further investigation is required to turn this information into real options to achieve treatments for ALMS patients.
Acknowledgments
The authors are thankful to the Registro Español de los Síndromes de Wolfram, Bardet–Biedl y Alström (REWBA), the European Union Rare Diseases Registry for Wolfram syndrome, Alström syndrome, Bardet–Biedl syndrome, and other rare diabetes syndromes (EURO-WABB) and BIOCAPS Project (from European Commission under the 7th Framework Programme, FP-7-REGPOT 2012-2013-1, grant agreement no FP7-316265). This work was supported by grants from Fondo de Investigación Sanitaria del Instituto de Salud Carlos III-FEDER (PI12/01853). María Álvarez-Satta and Sheila Castro-Sánchez are recipients of FPU fellowships from the Spanish Ministry of Education, Culture and Sports.
Disclosure
The authors report no conflicts of interest in this work.
References
Alström CH, Hallgren B, Nilsson LB, Asander H. Retinal degeneration combined with obesity, diabetes mellitus and neurogenous deafness: a specific syndrome (not hitherto described) distinct from the Laurence–Moon–Bardet–Biedl syndrome: a clinical, endocrinological and genetic examination based on a large pedigree. Acta Psychiatr Neurol Scand Suppl. 1959;(129):1–35. | |
Orphanet: an online rare disease and orphan drug data base [homepage on the Internet]. Paris: INSERM; 1997 [updated Nov 2012]. Available from: http://www.orpha.net/. Accessed March 13, 2015. | |
Titomanlio L, De Brasi D, Buoninconti A, et al. Alström syndrome: intrafamilial phenotypic variability in sibs with a novel nonsense mutation of the ALMS1 gene. Clin Genet. 2004;65(2):156–157. | |
Mahamid J, Lorber A, Horovitz Y, et al. Extreme clinical variability of dilated cardiomyopathy in two siblings with Alström syndrome. Pediatr Cardiol. 2013;34(2):455–458. | |
Marshall JD, Maffei P, Collin GB, Naggert JK. Alström syndrome: genetics and clinical overview. Curr Genomics. 2011;12(3):225–235. | |
Russell-Eggitt IM, Clayton PT, Coffey R, Kriss A, Taylor DS, Taylor JF. Alström syndrome. Report of 22 cases and literature review. Ophthalmology. 1998;105(7):1274–1280. | |
Marshall JD, Bronson RT, Collin GB, et al. New Alström syndrome phenotypes based on the evaluation of 182 cases. Arch Intern Med. 2005;165(6):675–683. | |
Satman I, Yilmaz MT, Gursoy N, et al. Evaluation of insulin resistant diabetes mellitus in Alström syndrome: a long-term prospective follow-up of three siblings. Diabetes Res Clin Pract. 2002;56(3):189–196. | |
Paisey RB, Paisey RM, Thomson MP, et al. Protection from clinical peripheral sensory neuropathy in Alström syndrome in contrast to early-onset type 2 diabetes. Diabetes Care. 2009;32(3):462–464. | |
Wu WC, Chen SC, Dia CY, et al. Alström syndrome with acute pancreatitis: a case report. Kaohsiung J Med Sci. 2003;19(7):358–361. | |
Paisey RB, Carey CM, Bower L, et al. Hypertriglyceridaemia in Alström’s syndrome: causes and associations in 37 cases. Clin Endocrinol (Oxf). 2004;(60):228–231. | |
Marshall JD, Beck S, Maffei P, Naggert JK. Alström syndrome. Eur J Hum Genet. 2007;15(12):1193–1202. | |
Maffei P, Boschetti M, Marshall JD, et al. Characterization of the IGF system in 15 patients with Alström syndrome. Clin Endocrinol (Oxf). 2007;66(2):269–275. | |
Mihai CM, Catrinoiu D, Toringhibel M, Stoicescu RM, Ticuta NP, Anca H. Impaired IGF1-GH axis and new therapeutic options in Alström syndrome patients: a case series. Cases J. 2009;2(1):19. | |
Romano S, Maffei P, Bettini V, et al. Alström syndrome is associated with short stature and reduced GH reserve. Clin Endocrinol. 2013;79(4):529–536. | |
Worthley MI, Zeitz CJ. Case of Alström syndrome with late presentation dilated cardiomyopathy. Intern Med J. 2001;31(9):569–570. | |
Hoffman JD, Jacobson Z, Young TL, Marshall JD, Kaplan P. Familial variable expression of dilated cardiomyopathy in Alström syndrome: a report of four sibs. Am J Med Genet A. 2005;135(1):96–98. | |
Zulato E, Favaretto F, Veronese C, et al. ALMS1-deficient fibroblasts over-express extra-cellular matrix components, display cell cycle delay and are resistant to apoptosis. PLoS One. 2011;6(4):e19081. | |
Joy T, Cao H, Black G, et al. Alström syndrome (OMIM 203800): a case report and literature review. Orphanet J Rare Dis. 2007;21(2):49. | |
Casey J, McGettigan P, Brosnahan D, et al. Atypical Alström syndrome with novel ALMS1 mutations precluded by current diagnostic criteria. Eur J Med Genet. 2014;57(2–3):55–59. | |
Shenje LT, Andersen P, Halushka MK, et al. Mutations in Alström protein impair terminal differentiation of cardiomyocytes. Nat Commun. 2014;(5):3416. | |
Louw JL, Corveleyn A, Jia Y, et al. Homozygous loss-of-function mutation in ALMS1 causes the lethal disorder mitogenic cardiomyopathy in two siblings. Eur J Med Genet. 2014;57(9):532–535. | |
Ozantürk A, Marshall JD, Collin GB, et al. The phenotypic and molecular genetic spectrum of Alström syndrome in 44 Turkish kindreds and a literature review of Alström syndrome in Turkey. J Hum Genet. 2015; 60(1):1–9. | |
NCBI: The National Center for Biotechnology information [homepage on the Internet]. Bethesda: US National Library of Medicine [updated Jan 2015]. Available from: http://www.ncbi.nlm.nih.gov/protein. Accessed March 13, 2015. | |
Marshall JD, Muller J, Collin GB, et al. Alström syndrome: mutation spectrum of ALMS1. Hum Mutat. 2015;36(7):660–668. | |
Waters AM, Beales PL. Ciliopathies: an expanding disease spectrum. Pediatr Nephrol. 2011;26(7):1039–1056. | |
Marshall JD, Maffei P, Beck S, Barrett TG, Paisey R, Naggert JK. Clinical utility gene card for: Alström syndrome – update 2013. Eur J Hum Genet. 2013;21(11). | |
Pereiro I, Hoskins BE, Marshall JD, et al. Arrayed primer extension technology simplifies mutation detection in Bardet–Biedl and Alström syndrome. Eur J Hum Genet. 2011;19(4):485–488. | |
Katagiri S, Yoshitake K, Akahori M, et al. Whole-exome sequencing identifies a novel ALMS1 mutation (p.Q2051X) in two Japanese brothers with Alström syndrome. Mol Vis. 2013;24(19):2393–2406. | |
Long PA, Evans JM, Olson TM. Exome sequencing establishes diagnosis of Alström syndrome in an infant presenting with non-syndromic dilated cardiomyopathy. Am J Med Genet A. 2015;167(4):886–890. | |
Wang X, Wang H, Cao M, et al. Whole-exome sequencing identifies ALMS1, IQCB1, CNGA3, and MYO7A mutations in patients with Leber congenital amaurosis. Hum Mutat. 2011;32(12):1450–1459. | |
McIntyre JC, Williams CL, Martens JR. Smelling the roses and seeing the light: gene therapy for ciliopathies. Trends Biotechnol. 2013;31(6):355–363. | |
Patel V, Chowdhury R, Igarashi P. Advances in the pathogenesis and treatment of polycystic kidney disease. Curr Opin Nephrol Hypertens. 2009;18(2):99–106. | |
Chang MY, Ong AC. Mechanism-based therapeutics for autosomal dominant polycystic kidney disease: recent progress and future prospects. Nephron Clin Pract. 2012;120(1):c25–c34. | |
Mockel A, Obringer C, Hakvoort TB, et al. Pharmacological modulation of the retinal unfolded protein response in Bardet–Biedl syndrome reduces apoptosis and preserves light detection ability. J Biol Chem. 2012;287(44):37483–37494. | |
Collin GB, Marshall JD, Ikeda A, et al. Mutations in ALMS1 cause obesity, type 2 diabetes and neurosensory degeneration in Alstrom syndrome. Nat Genet. 2002;31(1):74–78. | |
Hearn T, Spalluto C, Phillips VJ, et al. Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes. 2005;54(5):1581–1587. | |
Knorz VJ, Spalluto C, Lessard M, et al. Centriolar association of ALMS1 and likely centrosomal functions of the ALMS motif- containing proteins C10orf90 and KIAA1731. Mol Biol Cell. 2010; 21(21):3617–3629. | |
Heydet D, Chen LX, Larter CZ, et al. A truncating mutation of Alms1 reduces the number of hypothalamic neuronal cilia in obese mice. Dev Neurobiol. 2013;73(1):1–13. | |
Favaretto F, Milan G, Collin GB, et al. GLUT4 defects in adipose tissue are early signs of metabolic alterations in Alms1GT/GT, a mouse model for obesity and insulin resistance. PLoS One. 2014;9(10):e109540. | |
Collin GB, Marshall JD, King BL, et al. The Alström syndrome protein, ALMS1, interacts with α-actinin and components of the endosome recycling pathway. PLoS One. 2012;7(5):e37925. | |
Talior-Volodarsky I, Randhawa VK, Zaid H, Klip A. Alpha-actinin-4 is selectively required for insulin-induced GLUT4 translocation. J Biol Chem. 2008;283(37):25115–25123. | |
Satir P, Pedersen LB, Christensen ST. The primary cilium at a glance. J Cell Sci. 2010;123(4):499–503. | |
Christensen ST, Pedersen SF, Satir P, Veland IR, Schneider L. The primary cilium coordinates signaling pathways in cell cycle control and migration during development and tissue repair. Curr Top Dev Biol. 2008;(85):261–301. | |
Christensen ST, Clement CA, Satir P, Pedersen LB. Primary cilia and coordination of receptor tyrosine kinase (RTK) signalling. J Pathol. 2012;226(2):172–184. | |
Cardenas-Rodriguez M, Badano JL. Ciliary biology: understanding the cellular and genetic basis of human ciliopathies. Am J Med Genet C Semin Med Genet. 2009;151C(4):263–280. | |
Zaghloul NA, Katsanis N. Mechanistic insights into Bardet–Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119(3):428–437. | |
Marion V, Stutzmann F, Gérard M, et al. Exome sequencing identifies mutations in LZTFL1, a BBSome and smoothened trafficking regulator, in a family with Bardet–Biedl syndrome with situs inversus and insertional polydactyly. J Med Genet. 2012;49(5):317–321. | |
Jagger D, Collin G, Kelly J, et al. Alström syndrome protein ALMS1 localizes to basal bodies of cochlear hair cells and regulates cilium-dependent planar cell polarity. Hum Mol Genet. 2011;20(3):466–481. | |
Leitch CC, Lodh S, Prieto-Echagüe V, Badano JL, Zaghloul NA. Basal body proteins regulate Notch signaling via endosomal trafficking. J Cell Sci. 2014;(127):2407–2419. | |
Romano S, Milan G, Veronese C, et al. Regulation of Alström syndrome gene expression during adipogenesis and its relationship with fat cell insulin sensitivity. Int J Mol Med. 2008;21(6):731–736. | |
Huang-Doran I, Semple RK. Knockdown of the Alström syndrome-associated gene Alms1 in 3T3-L1 preadipocytes impairs adipogenesis but has no effect on cell-autonomous insulin action. Int J Obes (Lond). 2010;34(10):1554–1558. | |
Marion V, Stoetzel C, Schlicht D, et al. Transient ciliogenesis involving Bardet–Biedl syndrome proteins is a fundamental characteristic of adipogenic differentiation. Proc Natl Acad Sci U S A. 2009; 106(6):1820–1825. | |
Butler MG, Wang K, Marshall JD, et al. Coding and noncoding expression patterns associated with rare obesity-related disorders: Prader–Willi and Alström syndromes. Adv Genomics Genet. 2015;(5):53–75. | |
Woo YM, Bae J-B, Oh Y-H, et al. Genome-wide methylation profiling of ADPKD identified epigenetically regulated genes associated with renal cyst development. Hum Genet. 2014;133(3):281–297. | |
Chhin B, Negre D, Merrot O, et al. Ciliary beating recovery in deficient human airway epithelial cells after lentivirus ex vivo gene therapy. PLoS Genet. 2009;5(3):e1000422. doi:10.1371/journal.pgen.1000422. | |
McIntyre JC, Davis EE, Joiner A, et al. Gene therapy rescues cilia defects and restores olfactory function in a mammalian ciliopathy model. Nat Med. 2012;18(9):1423–1428. | |
Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet– Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci U S A. 2008;105(11):4242–4246. | |
Simons DL, Boye SL, Hauswirth WW, Wu SM. Gene therapy prevents photoreceptor death and preserves retinal function in a Bardet–Biedl syndrome mouse model. Proc Natl Acad Sci U S A. 2011;108(15):6276–6281. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
