Back to Journals » OncoTargets and Therapy » Volume 15
Airway Fusobacterium is Associated with Poor Response to Immunotherapy in Lung Cancer
Authors Chu S, Cheng Z, Yin Z, Xu J, Wu F, Jin Y, Yang G
Received 25 November 2021
Accepted for publication 15 February 2022
Published 28 February 2022 Volume 2022:15 Pages 201—213
DOI https://doi.org/10.2147/OTT.S348382
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Shujuan Chu,1 Zaixing Cheng,2 Zhongyuan Yin,3 Juanjuan Xu,4 Feng Wu,4 Yang Jin,4 Guanghai Yang2
1Department of Anesthesiology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, People’s Republic of China; 2Department of Thoracic Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, People’s Republic of China; 3Cancer Center, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, People’s Republic of China; 4Department of Respiratory Medicine, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, People’s Republic of China
Correspondence: Guanghai Yang, Department of Thoracic Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, People’s Republic of China, Tel +86 27 85351615, Email [email protected]
Purpose: There is a major limitation in the immunotherapy for solid cancer is that it only benefited a minority of cancer patients. This study aims to investigate whether the differential composition of the lung microbiome could affect the sustained clinical responses in lung cancers treated with immunotherapy.
Methods: Twenty-seven non-responders and 19 responders treated with anti-PD-1 therapy were included in the discovery set. Bacterial load in bronchoalveolar lavage from lung cancer patients was examined by quantitative PCR of 16S rRNA copies. Bacterial 16S rDNA was sequenced using the Illumina HiSeq on the 16S rDNA V3-V4 variable region. Operational taxonomic unit (OTU) analysis was performed using VSEARCH v2. The α-diversity and β-diversity were calculated using QIIME software.
Results: The mean copy number of bacterial 16S DNA levels significantly decreased after anti-PD-1 treatment (after: 1.8 ± 0.6× 104 copies per milliliter vs prior to treatment: 3.3 ± 1.1x104, p = 0.0036). In addition, longitudinal analysis revealed that microbial diversity was reduced taxonomically after treatment compared to those prior to the treatment (Shannon values: before: 3.291 ± 0.067 vs after: 2.668 ± 0.168, p < 0.01). Further, we observed a reduction of Fusobacterium nucleatum, including phylum Fusobacteria (p < 0.01), class Fusobacteria (p < 0.01), order Fusobacteria (p < 0.01), family Fusobacteria (p < 0.01), genus Fusobacteria (p = 0.025) in the responders post anti-PD-1 treatment. However, there was no significant difference of Fusobacterium in non-responders. An independent cohort was used to validate the levels of Fusobacterium, demonstrating that patients with higher abundance of Fusobacterium prior to treatment were significantly more likely to have poor response to anti-PD-1 therapy (p < 0.001).
Conclusion: Airway enriched Fusobacterium prior to anti-PD-1 therapy is associated with poor response in lung cancer, which indicated that potential resistance to immunotherapy can be attributed to lung microbiome.
Keywords: Fusobacterium, response, anti-PD-1 therapy, lung cancers
Introduction
In recent years, oncologic therapy using immune checkpoint inhibitors (ICIs) has led to great progress. The application of anti-programmed cell death-1/anti-programmed death ligand-1 (anti-PD-(L)1)/anti-cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) drugs has gradually become an indispensable treatment regimen for a variety of malignant tumors (such as non-small cell lung cancer (NSCLC) and renal cell carcinoma), and the survival time of patients with malignant tumors has been significantly prolonged.1 However, the rate of efficacy for ICIs is not high, and the responses of different patients to treatment also vary. Therefore, an increasing number of studies have focused on the effectiveness of immunotherapy, including the prediction of its efficacy and the alleviation of drug resistance. Some studies have suggested that PD-L1 expression on the surface of tumor cells, the tumor mutation burden, and the composition and structure of gut microbiota are potential biomarkers for immunotherapy.2
Lung cancer is one of the most common malignant tumors in China, with the highest mortality rate among all malignant tumors.3 Most patients with lung cancer have locally advanced or have extensive metastasis at the time of diagnosis; their 5-year survival rate is below 20%, and the overall prognosis is very poor.4,5 Previously, the lungs were considered sterile, but existing technology can detect microorganisms in the alveolar lavage fluid and lung tissues of normal people, potentially due to the migration of oropharyngeal flora and air inhalation.6 The results of these studies confirmed that the lower airway is not absolutely sterile. Other studies7,8 have shown not only that microorganisms exist in the lungs but also that differences exist in the microenvironment in the lungs, such as oxygen tension, pH value, and immune status, and that these differences may cause changes in the microbial spectrum in the lungs. Under normal circumstances, the microbial flora can maintain a healthy balance. However, once microbiota dysbiosis occurs under the action of certain factors, such disruptions can affect the local microenvironment, thereby affecting normal cell metabolism, leading to increased susceptibility to diseases and eventually the occurrence of malignant tumors. In a study by Peters et al,9 for the first time, a correlation between the respiratory microbiota and the prognosis of patients with NSCLC was established. They further showed that after 6 months of exposure to Haemophilus parainfluenzae, the number of inflammatory cells in the lungs increased,9 which promoted the occurrence of lung tumors in mice by promoting microangiogenesis and up regulating hypoxia-inducible factor 1-alpha (hif-1a) expression. Song et al10 revealed that microbe-induced T helper 17 (Th17) cells can promote lung cancer cell proliferation and angiogenesis. Tsay et al11 demonstrated that the microbiome regulated the immune response to cancer. In a mouse model, enterotoxigenic Bacteroides fragilis (ETBF) triggered the activation of signal transducer and activator of transcription 3 (STAT3) via Th17 cells to induce cancer through a Th17-dependent pathway. A study by Jin12 showed that the rapid increase in the number of bacteria in the lungs of mice with lung cancer leads to the secretion of interleukin 17 (IL-17) and IL-23 by gamma delta T cells (γδ T cells) and the release of inflammatory substances by neutrophils, which jointly promote tumor growth. Tumor volume has been shown to significantly decrease after receiving anti-infection therapy, suggesting that bacteria cause a violent immune response to stimulate tumor growth and that anti-infection treatment had antitumor effects. Other studies13,14 indicated that the progression-free survival (PFS) and overall survival (OS) of NSCLC patients receiving immunotherapy were worse than those who were not receiving anti-infection therapy. Liu et al15 found that the application of antibiotics may lead to the dysbiosis of Bacteroides and Veillonella in the gut, increasing the risk of immune-related reactions in lung cancer patients receiving anti-PD-1 antibody therapy. Perrone et al16 pointed out that since respiratory microbiome might influence pathogenesis, progression and outcome of lung cancer, a prospective study should be designed to evaluate airway microbiota changes in lung cancer, particularly patients under the treatment of immunotherapy. However, the impact of lung microbial compositions on the immunotherapy in lung cancers remains to be investigated. In this study, we proposed to decipher the relationship between the microbial composition in the lung local site prior to anti-PD-1 treatment and clinical response of the immunotherapy in patients with lung cancers. Furthermore, we sought to find the specific pathogens that are associated with resistance to the anti-PD-1 immunotherapy in lung cancers.
Materials and Methods
Subjects
Lung cancer patients who were treated with Nivolumab were selected in Union hospital Wuhan. All patients consented to an Institutional Review Board-approved study protocol for treatment and tissue collection. The inclusion criteria were as follows: (1) patients with a pathological diagnosis of lung cancer and no concurrent secondary tumor; (2) patients at clinical stage IV; (3) patients with measurable lesions; (4) patients with an expected survival time >6 months; (5) patients who had not received antitumor treatments such as surgery, chemotherapy, and radiotherapy. The exclusion criteria were as follows: (1) patients with brain metastases; (2) patients with autoimmune diseases; (3) Patients with anaplastic lymphoma kinase (ALK) or epidermal growth factor receptor (EGFR) mutations; and (4) patients with abnormal coagulation function. All patients signed an informed consent form. A total of 80 patients (43 males and 37 females) with advanced lung cancer were included in the study (discovery set and validation cohort). Sixty patients (75%) had an Eastern Cooperative Oncology Group (ECOG) score of ≤1; 67 patients had a history of smoking (84%). There were 34 cases of squamous carcinoma and 46 cases of adenocarcinoma; all were at stage IV. Twelve cases were combined with primary hypertension, 3 cases were combined with emphysema and pulmonary bullae, 4 cases were combined with type 2 diabetes, and 1 case was combined with chronic obstructive pulmonary disease, all without autoimmune diseases or other malignancies.
Medication Regimen
Anti-PD-1 monotherapy was used, and the recommended dose of 2 mg/kg or 3 mg/kg was prepared in the intravenous solution preparation center (PIVAS). All steps were conducted using strict aseptic techniques. The PD-1 antibody was added to 100 mL of normal saline. Intravenous infusion was performed using a sterile 0.2-μm filter infusion set for 30–60 min, followed by rinsing the tube with normal saline. One treatment cycle was approximately 2 to 3 weeks. During follow-up, the patients were treated with anti-PD-1 immunotherapy. The average follow-up time was 16.9 (3.6–38.9) weeks. Patients received anti-PD-1 during every follow-up visit, peripheral blood was collected before and after treatment for analysis, and changes in tumor volume were recorded by computed tomography. After 2 cycles of anti-PD-1 treatment, if the patients met the clinical assessment criteria, only changes in cellular immunity were observed. The primary endpoint of this study was the termination of follow-up, and the secondary endpoint was the patient’s choice to discontinue medication. Treatment efficacy was evaluated based on Response Evaluation Criteria in Solid Tumors 1.3 (RECIST1.3): complete remission (CR) refers to the complete disappearance of the lesions and no new lesions, with a duration >4 weeks; partial remission (PR) refers to a reduction in the longest diameter of the original lesion by >30%, with a duration >4 weeks; stable disease (SD) refers to a reduction in the longest diameter of the original lesions by <30% or an increase by <22%; and disease progression refers to an increase in the longest diameter of the primary lesions by >22% or the appearance of new lesions. Responders to ICIs include CR+PR.
Sampling and Microbial Sequencing
Bronchoscopy was performed before and after anti-PD-1 treatment. After local anesthesia, each patient was placed in the supine position, and a fiberoptic bronchoscope was inserted through the nasal cavity. After the lesions were examined, an appropriate amount of normal saline was injected into the lesion site to collect lavage fluid. One milliliter of lavage fluid stock solution was collected within 0.5 hours and stored in a −80°C refrigerator. Bacterial DNA was extracted using a PowerMax extraction kit (MoBio Laboratories) and stored in a −20°C freezer. The quantity and quality of DNA were determined using a NanoDrop 1000 spectrophotometer. After PCR amplification was performed using primers for the 16S rDNA V3-V4 variable region, AMPure beads were used to purify the amplification products. The purified products were subjected to NanoDrop detection, and the samples were mixed in equal amounts based on the measurements. The mixed samples were subjected to electrophoresis in a 2% agarose gel. The samples were recovered using an AXYGEN gel recovery kit. The recovered library was quantified using Qubit, and qPCR was performed to test the ligation efficiency. The actual concentration was calculated, followed by sequencing. Bacterial 16S rDNA was sequenced using the Illumina HiSeq, and the sequences of the 16S rDNA V3-V4 variable region of the bacterial flora were obtained. Operational taxonomic unit (OTU) analysis was performed using VSEARCH v2, 4, 4, including deduplication, clustering, and dechimerism. The sample sequences were clustered into OTUs based on 97% similarity, and the dilution curve tended to be flat, indicating that the sequencing depth was sufficient. The representative sequences of OTUs were selected using the default parameters, the representative sequences were annotated using VSEARCH based on the SILVA128 database to further generate the OTU list, and the microbial community composition of each sample was statistically analyzed at each taxonomic level. The abundance and classification of all OTUs in each sample were recorded, and the OTUs with a level of less than 0.001% of the total sequence in the sample were removed. The α-diversity and β-diversity indices at the OTU level were calculated using QIIME software. α-diversity, including the Chao1, Shannon, and Simpson indices, was calculated. Rank abundance curves, dilution curves, and box plots of the differences in the α-diversity index between groups were plotted to compare the abundance and evenness of the microbiota between the samples. β diversity analysis was conducted using principal coordinate analysis (PCoA) based on the weighted UniFrac distance to show the differences in the microbial community structure between different groups.
Quantitative PCR of 16S rDNA Copies and Fusobacterium Abundance
Bacterial load of BAL samples was evaluated on 16S rDNA copy number content using real-time quantitative PCR (qPCR) on BioRad RT-PCR Detection Systems using the following protocol: 98°C for 10 min, followed by 40 cycles of 96°C for 15 sec, and 60°C for 60 sec. The primers consisted of 1406F (5’-GYACWCACCGCCCGT-3’) and 1525R (5’-AAGGAGGTGWTCCARCC-3’). The qPCR was set up using 5µL of 2 x SYBR PCR Master mix (Applied Biosystems by Life Technologies), 2 µL of microbial template DNA and 1µL of primer mix. Bacterial numbers were normalized to the single copy gene Rpp4017 after a correction factor for bacterial numbers in the negative control samples. A serial dilution of DNA from E.coli was used to generate a standard curve.18 Quantitative PCR of Fusobacterium was performed to quantify the abundance of Fusobacterium species by using specific primers.19
Statistical Analysis
The data were processed using SPSS 23.3 statistical analysis software. Measurement data are presented as (X ± s) and were compared using the t-test; count data are presented as n (%) and were compared using the X2 test. p < 0.05 was considered statistically significant. PERMANOVA, using the R package “vegan”, was used to evaluate the markers of microbiota structure differentiation between groups. A Venn diagram was drawn based on the R package “VennDiagram” to visualize the shared and unique OTUs between groups. The Kruskal method in the R stats package was used to compare the differences in phylum, class, order, family, and genus among different taxonomic levels. For significant differences in the taxonomic levels of the microbiota, the difference between 2 groups was compared using the t-test or the Mann–Whitney U-test, and comparisons among 4 groups were performed using the Kruskal–Wallis test. All tests were performed using two-sided tests, and the test level (α) was 0.05.
Results
Patient Demographics
Eighty patients with advanced NSCLC were treated with anti-PD-1 immunotherapy and were followed during therapy until death. One year after the last inclusion, 19 patients (24%) were still on treatment, and 29 patients (36%) were still alive. Forty-four patients died within 6 months of inclusion and were categorized as poor responders, and 36 patients exhibited an OS longer than six months and were categorized as good responders. Out of the 80 subjects, 46 subjects (27 non-responders and 19 responders) were selected as the discovery set (Table 1). The median age [quartile] of responders in the discovery set was 64.0 [56.3, 75.0] years, 58.5 [55.3, 60.8] years in non-responders. Nine patients were female, and 10 patients were male in responder group from discovery set, 12 female patients and 15 male patients in non-responders. Body mass index (p = 0.895) and smoking status (p = 0.884) were not significantly different between responders and non-responders from discovery set. All the patients in discovery set showed an advanced stage IV. The remaining 34 subjects (17 non-responders and 17 responders) were used as the validation set and were not inspected until the analyses were performed (Table 1). The experimental design and analysis were included in the workflow, as shown in Figure 1.
 |
Table 1 Baseline Clinical Characteristics of the Study Population in the Discovery Set and Validation Set |
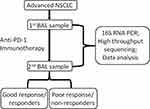 |
Figure 1 The workflow in this study. |
Lung Microbial Diversity Was Taxonomically Reduced After Anti-PD-1 Therapy
The bacterial quantification was firstly examined by qPCR and BAL from lung cancer patients contained 3.3 ± 1.1×104 copies of the 16S rRNA gene per mL (bacterial load). However, after anti-PD-1 treatment, the mean copy number of bacterial 16S DNA levels significantly decreased (1.8 ± 0.6x104, p = 0.0036) compared to those prior to treatment (Figure 2). Microbial community composition was further determined using Illumina MiSeq sequencing. Rarefaction analysis indicated that sequence depth was sufficient to obtain high sequence coverage (mean 97% and range 93–100%) (data not shown here). The total of 7 bacterial phyla was responsible for >99% of all sequence reads in each of the samples (Figure 3), including Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, Spirochaetes, Actinobacteria, and other unclassified bacteria. Shannon values and Simpson diversity index (1-D) were calculated to evaluate the microbial diversity in the various samples. The results shown that there were significantly higher Shannon values in the lungs before treatment compared to those post-anti-PD-1 therapy (before: 3.291 ± 0.067 vs after: 2.668 ± 0.168, p < 0.01), indicating that lungs after anti-PD-1 treatment exhibited a reduced species-richness community of microbiome compared to those before anti-PD-1 treatment. The Simpson index values ranged from 0.469 to 0.679 (0.599 ± 0.109) for the BAL samples before anti-PD-1 treatment, while there was a significant decrease of Simpson values (0.355 ± 0.088, p < 0.01) in BAL samples after immunotherapy, suggesting significantly lower diversity in the lung microbial composition after anti-PD-1 therapy. We next performed principal coordinate analysis (PCoA) to establish relationships between samples based on phylogenetic distance calculations. First, the OTUs found in 46 lung cancer patients in the discovery set were compared to each other based on before and after anti-PD-1 treatment. A weighted PCoA graph demonstrated that significant separation was observed from BALs before treatment and after treatment. In addition, the OTUs detected in first BAL samples before treatment were well separated from second BAL samples after treatment along principal coordinate 1, which accounted for 52% of the observed differences when we tested using weighted UniFrac analysis (Figure 4), which indicated that the diversity of microbial populations were significantly changed in advanced lung cancers after anti-PD-1 treatment compared to those prior to anti-PD-1 treatment. PC2 accounted for 11% variation and PC3 accounted for 7% variation of the microbial variation between BAL samples, indicating that there were other factors which would affect the microbiota changes.
Fusobacterium Was Associated with Poor Response Post Anti-PD-1 Therapy
We further sorted the OTUs to identify the genera present in all BAL samples of 46 lung cancer patients. At the top of the heatmap there were OTUs at Genus level that appeared to be present in all 92 BAL samples, which were retrieved from the data set and classified using RDP classifier and BLAST. Seven unique genera were found in 46 BAL samples collected prior to treatment and 46 BAL samples after the treatment of anti-PD-1: Rhodococcus (18.9%), Sphingobium (11.3%), Bacillus (2.3%), Escherichia (1.6%), Bergeyella (0.96%), Ralstonia (0.86%), Acinetobacter (0.91%), Klebsiella (0.57%), Actinomyces (0.30%), and Fusobacterium (0.39%) (Figure 5). To detect intra-individual changes in relative abundance and presence of microbes over time, we calculated a relative change matrix per taxonomical levels. From this relative change matrix per timeframe, we calculated the magnitude of microbial change using longitudinal regression analyses. We observed that the abundance of Actinobacteria (p < 0.01) at phylum level was increased over time upon anti-PD-1 treatment in responder group, while phylum Firmicutes (p < 0.01) and Phylum Fusobacteria (p < 0.01) were significantly reduced in responder group. Classes Actinobacteria (p < 0.01), Flavobacteria (p = 0.03) and Alpha proteobaceria (p < 0.01) increased after anti-PD-1 treatment in responder group. However, there was a decreased abundance of classes Bacilli (p < 0.01), Clostridia (p = 0.035), Fusobacteria (p < 0.01), and Gamma proteobacteria (p = 0.017) after anti-PD-1 treatment at class level when compared to those before treatment in responder group. Furthermore, we observed a reduction of Fusobacterium nucleatum, including Fusobacteria (p < 0.01) at order level, Fusobacteria (p < 0.01) at family level, Fusobacteria0 (p = 0.025) at the genus level (Figure 6), in the responders’ lungs following anti-PD-1 treatment. However, no significant change was found in each genera level of Fusobacterium in the non-responder group after anti-PD-1 treatment.
Among these microbial components, we were particularly interested in Fusobacterium because this genera has been reported to be predominantly represented in lungs20 and associated with immunotherapy in colorectal cancer and melanoma.14,21,22 To test the hypothesis that Fusobacterium nucleatum in cancer patients’ lungs prior to anti-PD-1 therapy may confer an increased risk of poor response of anti-PD-1 therapy, we collected an independent cohort of 17 responder patients compared with 17 non-responder patients. In BAL bacterial load (burden) analysis, there were no significant differences in total microbial biomass between responders and non-responders (Figure 7A). Bacterial levels of Fusobacterium were measured by quantitative PCR of its 16S ribosomal RNA gene. Results demonstrated that high abundance of Fusobacterium was significantly more likely to have poor response to anti-PD-1 therapy (Figure 7B). Taken together, our results strongly suggest that differential abundance of Fusobacterium prior to immunotherapy is associated with response effect in lung cancers.
Discussion
ICIs have become important therapeutic drugs for a variety of malignant tumors and can significantly improve the prognosis of cancer patients. However, a considerable number of patients cannot benefit from ICI treatment. An increasing number of studies have shown that microorganisms affect the efficacy of ICIs. Since the application of ICI antitumor therapy, the PFS of some patients with malignant tumors has been significantly prolonged. Frankel et al23 collected feces from melanoma patients treated with anti-PD-1 antibodies. The metagenomic sequencing results suggested that the relative abundance of Bacteroides caccae and Streptococcus parasanguinis in the intestinal flora of melanoma patients with a good anti-PD-1 response was high. For melanoma patients treated with different ICIs, the types of enriched microbes in intestinal flora corresponding to different therapeutic drugs were different when the treatment efficacy was similar. The relative abundance of Faecalibacterium prausnitzii was higher in the patients who received the combination of ipilimumab and nivolumab, while the patients who received pembrolizumab had a higher relative abundance of Dorea formicigenerans. In Gopalakrishnan’s study,21 melanoma patients treated with anti-PD-1 antibodies were divided into an effective group and an ineffective group based on clinical efficacy. The analysis of the intestinal flora of the 2 groups showed that there were differences in species richness, α or β diversity, and flora composition. Among them, in the intestinal flora of patients with a good response, the level of Faecalibacterium was higher, and the intestine of patients with a poor response had more Bacteroidales. Feces from the 2 groups of patients were transplanted into a mouse tumor model, and similar results were obtained. That is, the mice transplanted with the feces from the patients in the good response group had a lower tumor growth rate and had higher level of Faecalibacterium than did the mice transplanted with feces from the ineffective group. Studies have shown that if the intestinal flora of patients contains more Faecalibacterium, the level of CD4+CD8+ effector T cells in the peripheral circulation is higher, and that the high level of Bacteroidales will be accompanied by high levels of Treg cells and myeloid-derived suppressor cells. These cells play roles in immune regulation and suppression. Routy et al14 compared the differences in the efficacy of antibiotics for patients with 3 different types of tumors (NSCLC, renal cell carcinoma, and urothelial carcinoma), and all these patients received anti-PD-1 treatment. The PFS and OS of the patients who received antibiotics were lower than those who did not receive antibiotic treatment. The analysis of intestinal flora suggested that the level of Akkermansia bacteria in the intestine of patients who did not use antibiotics was higher than that of patients who used antibiotics. In addition, these 2 groups of patients were used as donors. Their feces were transplanted into the intestine of mice that had been pretreated with broad-spectrum antibiotics, and the mice were inoculated with MCA-205 sarcoma. The tumor growth rate in mice transplanted with feces from donors with a good response to anti-PD1 treatment was also slow, the level of C-X-C chemokine receptor type 4 (CXCR4)+ CD4+T cells in the peripheral circulation was high, and the expression of PD-L1 in splenic T lymphocytes was up regulated. The tumor growth rate in mice transplanted with feces from donors with a poor response to anti-PD-1 treatment was significantly faster than that in the other groups. When mice with a poor response were supplemented with Akkermansia, the efficacy of anti-PD-1 treatment was restored, and the response level improved in mice treated with broad-spectrum antibiotics. These results suggest that Akkermansia may play an important role in antitumor immunity. However, the mechanism by which some patients respond poorly or show drug resistance is not fully understood. These poor responses may be derived from the tumor or may be related to the patient’s own factors. For example, some tumor cells can directly upregulate the expression of PDL1 and bind to PD1, thereby producing an immunosuppressive effect. The type of HLA in the host and the structure of the intestinal flora also affect drug responses.14 Segmented filamentous bacteria can increase IL-22 levels in vivo through type 3 innate lymphoid cells,24 thereby regulating cell differentiation and changing the body’s response to anti-PD1 treatment. In our current study we have examined the microbial composition at local site of the lung in lung cancer patients with differential immunotherapeutic response. In addition, a substantial profile of Fusobacterium was higher in poor immunotherapeutic response. We further demonstrated that increased Actinobacteria (p<0.01), Flavobacteria (p=0.03) and Alpha proteobaceria (p<0.01); while reduced Bacilli (p<0.01), Clostridia (p=0.035), Fusobacteria (p<0.01), and Gamma proteobacteria (p=0.017) could also discriminate good and poor anti-PD-1 therapeutic response for advanced lung cancer treatment. These results would assist with potential in the immunotherapy of lung cancers and help monitoring or predicting disease progression during immunotherapy.
Our study showed that microbial diversity was reduced taxonomically after anti-PD-1 therapy compared to those prior to the treatment. Furthermore, we observed a reduction of Fusobacterium nucleatum (Fn), including Fusobacteria (p < 0.01) at phylum level, Fusobacteria (p < 0.01) at class level, Fusobacteria (p < 0.01) at order level, Fusobacteria (p < 0.01) at family level, Fusobacteria0 (p = 0.025) at the genus level, in the responders’ lungs following anti-PD-1 treatment. However, no significant change was found in each genera level of Fusobacterium in the non-responder group after anti-PD-1 treatment. Using an independent cohort of 17 responder patients compared with 17 non-responder patients, bacterial levels of Fusobacterium measured by quantitative PCR demonstrated that higher abundance of Fusobacterium was significantly more likely to have poor response to anti-PD-1 therapy. Therefore, airway enriched Fusobacterium prior to anti-PD-1 therapy was associated with poor response in advanced lung cancer. Fn belongs to the genus Fusobacterium and is a common gram-negative anaerobe. The abundance of Fn is affected by environmental factors and can be increased in smoking populations.25,26 Fn has a strong colonization ability and can invade oral epithelial cells and promote the release of proinflammatory cytokines.25,26 Fn was found to be enriched in intestinal tissues, encode and produce adhesion proteins such as Fusobacterium adhesin A (FadA), familial adenomatous polyposis-2 (Fap2), RadD, and Aid1, and can act on a variety of immune cells (endothelial cells, NK cells, monocytes, etc.) and proteins (immunoglobulin, Cadherin, etc.) to evade host defenses and induce abnormal host immune responses.27–29 Fn was found to be present in colon cancer as well as in primary tumors and liver metastases and that its presence is correlated with poor patient prognosis.30 Fn-positive colorectal tumors were transplanted in mice, and antibiotics delayed tumor growth.31 Further, they showed that Fn interacted with toll-like receptor (TLR4) to regulate autophagy, reduce apoptosis and induce chemotherapy tolerance.32 Fap2 is an adhesin that mediates Fn recognition and binding to colorectal cancer cells. Fap2 can bind to TIGHT receptors on natural killer cells and other tumor-infiltrating lymphocytes.33 The TIGHT pathway can inhibit the cytotoxic function of these cells, thereby protecting Fn and its nearby tumor cells from being killed by immune cells.34 In addition, Fn can also recruit bone marrow-derived immune cells to construct a microenvironment that is conducive to tumor growth and block the antitumor immune response of natural killer cells. Epidemiological investigations have shown that Fn can also cause gene mutations and genomic mutations in the body but that Fn does not encode toxins and its gene sequence almost completely lacks sequences that encode typical virulence factors.35 However, there is a limitation that our study lacked a reverse validation and the underlying mechanisms require further investigation.
In conclusion, we performed a metagenomic sequencing analysis on microbial compositions collected from local site of the lungs before and after anti-PD-1 treatment and further analyzed the relationship between bacterial load and diversity with the clinical response to anti-PD-1 immunotherapy. Importantly, we demonstrated that airway enriched Fusobacterium prior to anti-PD-1 therapy is associated with resistance to anti-PD-1 response in lung cancer, providing potential implication in treatment resistance in immunotherapy.
Ethics Approval and Consent to Participate
The study was performed in accordance with relevant guidelines and regulations, following the approval of the licensing committee of Tongji Medical College, Huazhong University of Science and Technology. Written informed consent was obtained from the patients. This work received approval from the institution ethics committee and conformed to the tenets of the Declaration of Helsinki.
Consent to Participate
All patients signed written informed consent before study entry.
Consent for Publication
Patients signed informed consent regarding the publication of their data.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study is supported in part by the National Natural Science Foundation of China (NSFC Grant No. 81800094 to J.X.) and in part by the National Natural Science Foundation of China (NSFC Grant No. 81973989 to G.Y.).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Homet Moreno B, Ribas A. Anti-programmed cell death protein-1/ligand-1 therapy in different cancers. Br J Cancer. 2015;112:1421–1427. doi:10.1038/bjc.2015.124
2. Otoshi T, Nagano T, Tachihara M, Nishimura Y. Possible biomarkers for cancer immunotherapy. Cancers. 2019;11. doi:10.3390/cancers11070935
3. Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer. 2019;10:3–7. doi:10.1111/1759-7714.12916
4. Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2–12. doi:10.3978/j.issn.1000-9604.2015.01.06
5. She J, Yang P, Hong Q, Bai C. Lung cancer in China: challenges and interventions. Chest. 2013;143:1117–1126. doi:10.1378/chest.11-2948
6. Dickson RP. The microbiome and critical illness. Lancet Respir Med. 2016;4:59–72. doi:10.1016/S2213-2600(15)00427-0
7. Faner R, Sibila O, Agusti A, et al. The microbiome in respiratory medicine: current challenges and future perspectives. Eur Respir J. 2017:49. doi:10.1183/13993003.02086-2016
8. Shukla SD, Budden KF, Neal R, Hansbro PM. Microbiome effects on immunity, health and disease in the lung. Clin Transl Immunol. 2017;6:e133. doi:10.1038/cti.2017.6
9. Peters BA, Hayes RB, Goparaju C, Reid C, Pass HI, Ahn J. The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol Biomarkers Prev. 2019;28:731–740. doi:10.1158/1055-9965.EPI-18-0966
10. Song L, Ma S, Chen L, Miao L, Tao M, Liu H. Long-term prognostic significance of interleukin-17-producing T cells in patients with non-small cell lung cancer. Cancer Sci. 2019;110:2100–2109. doi:10.1111/cas.14068
11. Tsay JJ, Wu BG, Badri MH, et al. Airway microbiota is associated with upregulation of the pi3k pathway in lung cancer. Am J Respir Crit Care Med. 2018;198:1188–1198. doi:10.1164/rccm.201710-2118OC
12. Jin C, Lagoudas GK, Zhao C, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176:998–1013 e16. doi:10.1016/j.cell.2018.12.040
13. Derosa L, Hellmann MD, Spaziano M, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. 2018;29:1437–1444. doi:10.1093/annonc/mdy103
14. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi:10.1126/science.aan3706
15. Liu T, Xiong Q, Li L, Hu Y. Intestinal microbiota predicts lung cancer patients at risk of immune-related diarrhea. Immunotherapy. 2019;11:385–396. doi:10.2217/imt-2018-0144
16. Perrone F, Belluomini L, Mazzotta M, et al. Exploring the role of respiratory microbiome in lung cancer: a systematic review. Crit Rev Oncol Hematol. 2021;164:103404. doi:10.1016/j.critrevonc.2021.103404
17. Doring G, Parameswaran IG, Murphy TF. Differential adaptation of microbial pathogens to airways of patients with cystic fibrosis and chronic obstructive pulmonary disease. FEMS Microbiol Rev. 2011;35:124–146. doi:10.1111/j.1574-6976.2010.00237.x
18. Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi:10.1371/journal.pone.0008578
19. McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:e53653. doi:10.1371/journal.pone.0053653
20. Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res. 2012;160:258–266. doi:10.1016/j.trsl.2012.02.005
21. Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi:10.1126/science.aan4236
22. Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi:10.1126/science.aao3290
23. Frankel AE, Coughlin LA, Kim J, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 2017;19:848–855. doi:10.1016/j.neo.2017.08.004
24. Sano T, Huang W, Hall JA, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163:381–393. doi:10.1016/j.cell.2015.08.061
25. Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17:156–166. doi:10.1038/s41579-018-0129-6
26. Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–147. doi:10.1016/j.mib.2014.11.013
27. Kaplan A, Kaplan CW, He X, McHardy I, Shi W, Lux R. Characterization of aid1, a novel gene involved in Fusobacterium nucleatum interspecies interactions. Microb Ecol. 2014;68:379–387. doi:10.1007/s00248-014-0400-y
28. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi:10.1016/j.chom.2013.07.012
29. Shokeen B, Park J, Duong E, et al. Role of FAD-I in fusobacterial interspecies interaction and biofilm formation. Microorganisms. 2020;8:70. doi:10.3390/microorganisms8010070
30. Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi:10.1126/science.aal5240
31. Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi:10.1136/gutjnl-2015-310101
32. Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–63 e16. doi:10.1016/j.cell.2017.07.008
33. Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi:10.1016/j.immuni.2015.01.010
34. Dougall WC, Kurtulus S, Smyth MJ, Anderson AC. TIGIT and CD96: new checkpoint receptor targets for cancer immunotherapy. Immunol Rev. 2017;276:112–120. doi:10.1111/imr.12518
35. Sanders BE, Umana A, Lemkul JA, Slade DJ. FusoPortal: an interactive repository of hybrid MinION-sequenced fusobacterium genomes improves gene identification and characterization. mSphere. 2018;3. doi:10.1128/mSphere.00228-18
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






