Back to Journals » Research and Reports in Urology » Volume 13
A Systematic Review of Artificial Intelligence in Prostate Cancer
Authors Van Booven DJ, Kuchakulla M, Pai R, Frech FS , Ramasahayam R, Reddy P , Parmar M, Ramasamy R, Arora H
Received 22 November 2020
Accepted for publication 11 January 2021
Published 22 January 2021 Volume 2021:13 Pages 31—39
DOI https://doi.org/10.2147/RRU.S268596
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jan Colli
Derek J Van Booven,1 Manish Kuchakulla,2 Raghav Pai,2 Fabio S Frech,2 Reshna Ramasahayam,2 Pritika Reddy,2 Madhumita Parmar,2 Ranjith Ramasamy,2,3 Himanshu Arora1– 3
1John P Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL, USA; 2Department of Urology, Miller School of Medicine, University of Miami, Miami, FL, USA; 3The Interdisciplinary Stem Cell Institute, Miller School of Medicine, University of Miami, Miami, FL, USA
Correspondence: Himanshu Arora Email [email protected]
Abstract: The diagnosis and management of prostate cancer involves the interpretation of data from multiple modalities to aid in decision making. Tools like PSA levels, MRI guided biopsies, genomic biomarkers, and Gleason grading are used to diagnose, risk stratify, and then monitor patients during respective follow-ups. Nevertheless, diagnosis tracking and subsequent risk stratification often lend itself to significant subjectivity. Artificial intelligence (AI) can allow clinicians to recognize difficult relationships and manage enormous data sets, which is a task that is both extraordinarily difficult and time consuming for humans. By using AI algorithms and reducing the level of subjectivity, it is possible to use fewer resources while improving the overall efficiency and accuracy in prostate cancer diagnosis and management. Thus, this systematic review focuses on analyzing advancements in AI-based artificial neural networks (ANN) and their current role in prostate cancer diagnosis and management.
Keywords: prostate cancer, active surveillance, clinical trials, artificial intelligence
Introduction
Prostate cancer is the most commonly diagnosed non-skin malignancy in men and second leading cause of mortality from cancer. Furthermore, it is estimated that 1 in 6 American men will be affected by this ailment during their lifetimes.1 Challenges faced during the course of prostate cancer management include increased volume of prostate biopsies and a shortage of urological pathologists, which puts a strain on diagnosis of prostate cancers.2 Additionally, the presence of a variability of pathological grading can result in an overtreatment or under treatment of prostate cancer.3
Artificial Intelligence (AI) and machine learning in the healthcare is a new area of research which has recently gained a lot of interest. AI often makes use of artificial neural networks (ANN) that use statistical models that are directly inspired by and partially modeled on biological neural networks. They are capable of modeling and processing nonlinear relationships between inputs and outputs in parallel. Figure 1 shows a schematic of the structure of an ANN. In prostate cancer, the use of AI overall has shown to be beneficial to aid in a standardized pathological grading to assess prostate cancer stratification and treatment. Additionally, AI shows promise in automating the assessment of characterization and severity of prostate cancer based on image-based tasks including in histopathologic, MRI, and biomarker diagnosis.4 Furthermore, certain patients that are diagnosed with prostate cancer which is thought to be more indolent can continue with repeated forms of surveillance including prostate biopsies, PSA, and other forms of digital testing through MRI or rectal examinations unless they experience any physiological side effects5. AI can help improve these forms of surveillance and will be amongst some of the essential tools to urological pathologists and to the field of urology as a whole as technology continues to improve and help patient prognosis over time. In this present study, we systematically reviewed the literature to determine how AI can be used to improve the diagnosis and management of prostate cancer.
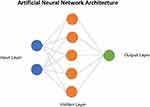 |
Figure 1 Schematic depicting the architecture of an artificial neural network. |
Materials and Methods
In this systematic review we used Pubmed, Embase, and Web of Science to identify and screen manuscripts pertaining to the use of machine learning and active surveillance in the diagnosis, staging, and treatment of prostate cancer. This review was conducted according to the Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA). The keywords used include: (((((((((((prostate cancer) OR prostatic cancer) OR guided prostate biops*) OR guided prostatic biops*) OR ultrasound guided prostate biops*) OR ultrasound guided prostatic biops*) OR prostate biops*) OR prostatic biops*) OR gleason scor*)) AND (((((artificial intelligence) OR deep learning) OR machine learning) OR artificial neural network) OR neural network)) AND (((clinical trial*) OR active surveillance) OR active surveillance trial). Preliminary results showed 341 manuscripts. After duplicate removal, an exclusion criteria was applied to 283 manuscripts. This exclusion criteria is as follows: obviously not specific to the topic in question, case report, review, opinion, conference paper, commentary, survey, animal studies. After exclusion criteria was implemented on title and abstract, 81 full-text manuscripts were assessed for eligibility. After full-text assessment of eligibility, 19 manuscripts will be included in this systematic review. Figure 2 shows a flow diagram depicting the flow of information through the different phases of the literature search. A summary of included studies is presented in Table 1.
 |
Table 1 Summary of All Articles Included in the Systematic Review |
 |
Figure 2 Flow diagram representing different phases of the systematic literature research according to PRISMA criteria. Notes: PRISMA figure adapted from Liberati A, Altman D, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Journal of clinical epidemiology. 2009;62(10). Creative Commons.43 |
Literature Review
Artificial Neural Network (ANN) and Prostate-Specific Antigens (PSA)
Identification of elevated PSA level is regarded as one of the most common clinical tool for diagnosis of prostate cancer.6 However, an elevated PSA level is present in several other benign prostatic conditions that can lead to unnecessary prognostic procedures.7 Generally, a PSA level greater than 4 ng/mL is considered for early detection of prostate cancer, but up to 20% of patients diagnosed with prostate cancer never reach that threshold.7 There has been a great need to diagnose prostate cancer in men with borderline PSA levels to improve health outcomes using new non-invasive methods.
Numerous studies have evaluated the utility of PSA and other clinical characteristics in the setting of an ANN in detecting prostate cancer and/or its progression. For example, Djavan et al developed an ANN to enhance the sensitivity and specificity of prostate cancer detection.8 The study consisted of 1246 men with the primary aim of predicting the presence of prostate cancer or benign prostatic tissue in men with serum total PSA levels from 2.5 to 4 ng/mL and from 4 to 10 ng/mL compared to conventional tools. At 95% sensitivity, the ANN model for patients with total PSA levels from 4 to 10 ng/mL produced specificity, positive predictive value, negative predictive value, and receiver operator characteristic (ROC) curve area under the curve values that were superior to any of the other comparative parameters.8 A different study by Stojadinovic et al looked at various factors and determined that PSA density was the most decisive variable and showed the decision tree providing a net benefit compared to a logistic regression model. In this retrospective study, the model had a potential to reduce unnecessary biopsies without missing significant diagnoses.9
Finne et al developed an ANN using free PSA with the goal of eliminating unnecessary prognostic testing in false-positive PSA results.10 The model was constructed using data on total PSA, the proportion of free PSA, digital rectal examination (DRE), and prostate volume from 656 men with total serum PSA concentrations of 4 to 10 ng/mL in Finland. Results showed that, at a 95% sensitivity level, 19% of the false-positive PSA results could be eliminated by using the proportion of free PSA versus 24% with the logistic regression model and 33% with the ANN model. These findings suggest that an ANN method has value to be incorporated into prostate cancer screening to decrease the number of false-positive results.10
Ge et al developed a logistic regression and ANN model to aid in diagnosis of prostate cancer. They utilized predictors including age, percent free PSA, prostate volume, and PSA density from 586 men with prostate cancer confirmed by biopsy to train their algorithms. While they found no significant difference between the two models, they concluded that both models have high diagnostic validity and have potential to be included in practice to prevent unnecessary biopsies.11
Stephan et al developed an ANN utilizing percentage of free PSA (%fPSA) to identify men at increased risk of a positive prostate biopsy for cancer.12 The study enrolled 1188 men with benign prostates or prostate cancer with input data that included total PSA, %fPSA, patient age, prostate volume, and digital rectal examination (DRE) status. The use of the ANN in men with 2–10 μg/L total PSA enhanced the specificity of %fPSA by 20–22%. Thus, the use of an %fPSA-based ANN is able to enhance detection accuracy over %fPSA alone and prevent unnecessary biopsies.12 Together, there are multiple studies which reflect the relevance of using artificial intelligence-based ANN tools which could be effectively integrated with some of the vital tools commonly used in prostate cancer detection.
Artificial Neural Network and Histopathologic Diagnosis of Prostate Cancer
The histopathological identification of prostatic adenocarcinoma is often essential for establishing a diagnosis of prostate cancer.13 However, sometimes histopathologic variability occurs due to inter- and intra-observer variability. This variability could impact the evaluation of biologic aggressiveness of prostate cancer and the identification of patients at high risk for progression.14 A solution to this has been to introduce an artificial intelligence interface to accurately detect, localize and grade histopathologic slides. Early studies such as Bhele et al focused on detecting the difference in histology slides between Gleason score. Gleason grading allows the stratification of the aggressiveness of prostate cancer into low (grade group 1), intermediate (grade group 2 and 3) or high-risk group (grade group 4 and 5). Group 1 is designated by Gleason score (GS) 6, group 2 by GS7 or 3+4, group 3 by GS7 or 4+3, group 4 by GS8, and group 5 by GS9-10, respectively. The authors focused on detecting the difference between GS3 and GS4.15 The model in this particular study was trained on 38 radical prostatectomy samples yielding 105 images. The tool reached a concordance on outlines of G3, G4, and benign epithelium of 67–81%. Thus, the AI model showed a good promise of being able to distinguish between 3+4 and 4+3 patients.
Using digitized slides from 1247 men, Ström et al trained ANNs to assess prostate biopsies.2 The networks were evaluated by predicting the presence, extent, and Gleason grade of malignant tissue. The AI tool achieved an area under the ROC curve of 0.997 for distinguishing between benign and malignant biopsy cores on the independent test dataset and 0.986 on the external validation dataset. These results were comparable to the accuracy of an international expert in prostate pathology.2 An AI system such as this can provide a standardized grading method and pathology expertise in underserved areas.
In another study by Lawrentschuk et al, a polychotomous logistic regression (PR) model and an ANN was developed for predicting the biopsy results using 3025 men undergoing biopsy with PSA <10ng/dL.16 Clinical predictors of age, PSA, abnormal digital rectal examination, positive transrectal ultrasound (TRUS) and prostate volume were also considered during the development. It was suggested that an inclusion of additional predictors may improve the performance of the models; however, the ANN was unable to distinguish between the four biopsy outcomes that were utilized in their validation. The authors suggest that a more tempered enthusiasm should be used with ANN and that great care must be used in training them properly for effective application.17
In yet another study by Remzi et al, an ANN was developed to predict the presence of prostate cancer and predict the outcome in repeat biopsies.18 This model was trained using variables such as age, PSA, f/t PSA ratio, digital rectal examination findings, PSA velocity, and the transrectal ultrasound-guided variables of prostate volume, transition zone volume, PSAD, and PSA-TZ from 820 men with PSA levels between 4 and 10 ng/mL. This ANN showed a strong pattern of prediction for prostate cancer in patients who had an initial negative biopsy. Overall, the ANN achieved a 68% specificity and 95% sensitivity.18 Together, there are multiple studies which reflects the potential of AI ANNs to allow more accurate counseling of patients and overcome histopathologic variabilities.
Artificial Neural Network and Magnetic Resonance Imaging (MRI) Diagnosis of Prostate Cancer
Magnetic resonance imaging (MRI) has been investigated as a modality for prostate cancer detection and determination of aggressiveness.19,20 MR spectroscopic T2-weighted MR imaging, and apparent diffusion coefficient (ADC) have been valuable tools to assess prostate cancer, but their use still lacks consensus.21 Using these tools, expert users can easily detect malignant tumors, but determining their aggressiveness using MRI with good reliability is more difficult. Thus, automatic classification using machine learning has been proposed as a solution to provide more accurate and consistent results to aid clinicians’ management.21 In the similar context, Fehr et al proposed a technique of utilizing machine learning-based automatic classification of prostate cancer aggressiveness by combining apparent diffusion coefficient (ADC) and T2-weighted MRI-based texture features.21 This technique was able to differentiate between the high GS (>7) and low GS6, as well as between 7 (3+4) and 7 (4+3) cancers. Furthermore, this model could distinguish between high and low Gleason grades with 93% accuracy for cancers occurring in both peripheral (PZ) and transition (TZ) zones and 92% for cancers occurring in the PZ alone.21 These results were far superior to using only ADC and suggest that this method can help provide reasonably accurate classification of Gleason patterns.
In addition, in another study by Antonelli et al, the authors used quantitative MRI and clinical features from 164 men to construct machine learning classifiers.22 Following model validation, these classifiers were able to predict Gleason 4 in prostate tumors with greater accuracy than the three board-certified radiologists which participated in the study. It was suggested that these AI classifier tools could be useful to non-invasively detect the progression of tumors and aid in decisions regarding active surveillance programs.22
In yet another study by Toivonen et al, the authors developed machine learning tools to predict prostate cancer aggressiveness by using optimized high-quality MRI data sets.23 A classifier system was created based on multiple texture features of high-quality T2 weighted images, DWI (diffusion weighted imaging), and T2 relaxation maps from 100 patients for prediction of PCa Gleason score dichotomized as 3+3 (low risk) vs >3+3 (high risk). Results suggested that texture feature analysis of DWI, post-processed using monoexponential and kurtosis models, and T2w demonstrated good classification performance for Gleason score of prostate cancer.23 Together, these are multiple studies which reflects the potential of AI ANNs to allow more effective integration with modern surveillance tools such as MRI to effectively help in prostate cancer surveillance.
Artificial Neural Network in Biomarker Diagnosis and Risk Stratification
Diagnosis and prognosis of prostate cancer has been guided by PSA level testing. However, concerns about absolute accuracy can sometimes lead to patients being given invasive treatment options when active surveillance might provide better outcomes in these men.24 Over the past 10 years, an avalanche of biomarkers have been identified and are put into clinical assays.25–34 While several of these biomarkers have been studied and characterized, there is no standard overlap between all of these assays due to the function of each assay, and there is no absolutely perfect list of biomarkers to look at when predicting diagnosis and prognosis. Thus, it is important to be able to identify and evaluate any new biomarkers for their clinical significance in a way that is both meaningful and accurate. Therefore, ANNs can play an instrumental role in analyzing and validating the biomarkers. For example, one study suggested that Ki67 is an important marker of survival and disease progression.35 To confirm this, Green et al created an ANN that was designed to validate Ki67 gene expression while comparing it to another potential candidate in DLX2.36 Univariate analysis showed that both Ki67 and DLX2 were significant in predictiveness of future metastases. Nevertheless, only 6.8% of prostate cancer patients have high expression of Ki67. Thus, this study showed that these 2 biomarkers could be used to identify candidates for targeted therapy only.36
In addition to gene expression, proteomics can also be useful when identifying potential biomarkers. For example, Kim et al developed a novel approach by combining targeted proteomics with computational biology to discover new potential proteomic signatures for prostate cancer.37 The study started with 133 differentially expressed proteins that were evaluated with synthetic peptides in a 74-patient cohort. Then they applied machine learning approaches to develop clinical predictive models using these candidates. Results showed that computationally guided proteomics can be used to discover novel non-invasive biomarkers. Overall, there are numerous studies which reflect the potential of AI ANNs to allow more effective identification and validation of biomarkers to aid in prostate cancer surveillance.
Artificial Neural Network in Patient Interaction and Patient-Centered Treatment
Patients diagnosed with prostate cancer often remain confused as to the treatment options that are available to them. Thus, understanding how certain therapies are put into place may allow for ease of mind and improvements in patient satisfaction. Auffenberg et al developed a registry that used ANN to better allow patients to take charge of their care. This registry, which is called askMUSIC, takes data from 45 urology practices within the Michigan Urological Surgery Improvement Collaborative (MUSIC).38 This registry data is used to create a random forest machine learning model which could predict prostate cancer treatment options. Patients can go to askMUSIC website and interact with the registry data and predicted treatment to show therapy options to alleviate fear about a given therapy from the patient perspective.
In the similar context, the PRODIGE project by Alitto et al uses an Umbrella Protocol which focuses on standardization of data sharing.39 Within this protocol, a standardized knowledge sharing process is implemented using semi-formal ontology representing clinical variables. This process can be adapted to use with machine learning or traditional statistics. The standardization of these techniques supports the multifactorial decision support systems (DSS) which can be seen as the basis for future patient-level support therapy decisions. Together, there are multiple studies which reflect the potential of AI ANNs to allow the development of effective patient-centric tools to help with educating the patients about the treatment options and disease progression/regression.
Artificial Neural Network in Creating a Classification System for Prostate Cancer Risk Stratification
Current risk stratification as defined by the National Comprehensive Cancer Network (NCNN) guidelines relies on TNM (tumor, node, metastasis) score, Gleason grade, PSA level, and biopsy results.5 These factors guide patients into risk groups which are stratified into 7 categories ranging from very low to very high. Based on these risk categories, physicians can guide patients into various personalized treatment options ranging from active surveillance, radical prostatectomy, radiation, and hormone therapy. While these risk categories are a good basis for stratification, none of the risk categories looks at the potential for recurrence. Thus, a better understanding of mechanisms behind recurrence could educate us to adjust the risk factors and therefore make the right choices for therapy.
Kumar et al focused on this recurrence prediction by creating 2 different convolutional neural networks (CNN) analyzing H&E images.40 The first CNN detected individual nuclei while the second CNN classified the patches around the nuclear centers. There was a voting process which yielded probabilities of recurrence from the patients. The model was trained on 80 case/control pairs and then validated on 30 recurrent and 30 non-recurrent controls. The end yield was 0.81 AUC for accuracy. Thus, providing a deep learning tool that could look at recurrence in conjunction with other risk factors to come up with a new scoring system that could be more accurate than currently used.
In addition to recurrence, the current standards in identifying risk factors for prostate cancer often fails to take basic demographics into account. Nevertheless, a study by Naguib et al evaluated a combined approach of ANN and traditionally statistics known about various risk factors.41 The neural network in this study was trained on conventional factors like age, stage, bone scan findings, grade, PSA and treatment along with 2 experimental markers of immunostaining for bcl-2 and p53. By considering conventional risk factors alone, the prediction models achieved only 60% accuracy in identifying the patients with condition. However, this efficiency was improved to 80% when the experimental markers were included. Thus, there was a significant improvement in accuracy of the model with an increase in the amount of data fed to the algorithm. This study also underscored the importance of including experimental markers for testing into building the networks as each piece of data could impact the model accuracy profoundly. Overall, there are multiple studies which reflect the potential of AI ANNs to allow the development of effective classification system for prostate cancer risk stratification.
Conclusion
The use of artificial intelligence to manage medical problems has been discussed for quite some time. Nevertheless, it is only recently that technological advances have allowed us to make significant strides. AI can allow us to recognize difficult relationships and manage enormous data sets, which is a task that is both extraordinarily difficult and time consuming for humans. Moreover, the diagnosis and subsequent risk stratification that is required for active surveillance trials lends itself to enormous subjectivity. By using AI algorithms and reducing the level of subjectivity, it is possible to use fewer resources while improving the overall efficiency and accuracy in trials.
The economic burden of the management of localized prostate cancer must be acknowledged as well. The 10-year costs of managing low- and high-risk patients with prostate cancer have been estimated to be $45,957 and $188,928, respectively. However, a broader incorporation of AI can certainly help in mitigating this burden.42 Furthermore, AI makes it feasible to accomplish this while maintaining or perhaps even improving current outcomes in active surveillance trials. Thus, it is essential to not only acknowledge but continue to improve the current pipelines to reduce costs, improve outcomes, and drive the future of urologic oncology forward as a whole.
Acknowledgments
We thank all the mentors (Dr. Dipen J Parekh, Dr Joshua M Hare) for their insights, suggestions, and support during this study. Additionally, we would like to thank the American Urological Association Research Scholar Award and Stanley Glaser Award to HA and RR.
Disclosure
The authors report no conflicts of interest for this work. Declaration of interests: the authors declare no competing interests.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi:10.3322/caac.21262
2. Strom P, Kartasalo K, Olsson H, et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based. Diagnostic Study, Lancet Oncol. 2020;21:222–232. doi:10.1016/S1470-2045(19)30738-7
3. Harmon SA, Tuncer S, Sanford T, Choyke PL, Türkbey B. Artificial intelligence at the intersection of pathology and radiology in prostate cancer. Diagn Interv Radiol. 2019;25:183–188.
4. Harmon SA, Tuncer S, Sanford T, Choyke PL, Turkbey B. Artificial intelligence at the intersection of pathology and radiology in prostate cancer. Diagn Interv Radiol. 2019;25:183–188.
5. Carroll PH, Mohler JL. NCCN guidelines updates: prostate cancer and prostate cancer early detection. J Natl Compr Canc Netw. 2018;16:620–623. doi:10.6004/jnccn.2018.0036
6. Partin AW, Oesterling JE. The clinical usefulness of prostate specific antigen: update 1994. J Urol. 1994;152:1358–1368. doi:10.1016/S0022-5347(17)32422-9
7. Catalona WJ, Smith DS, Ratliff TL, et al. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N Engl J Med. 1991;324:1156–1161. doi:10.1056/NEJM199104253241702
8. Djavan B, Remzi M, Zlotta A, Seitz C, Snow P, Marberger M. Novel artificial neural network for early detection of prostate cancer. J Clin Oncol. 2002;20:921–929. doi:10.1200/JCO.2002.20.4.921
9. Stojadinovic M, Stojadinovic M, Pantic D. Decision tree analysis for prostate cancer prediction. Srp Arh Celok Lek. 2018;147:39.
10. Finne P, Finne R, Auvinen A, et al. Predicting the outcome of prostate biopsy in screen-positive men by a multilayer perceptron network. Urology. 2000;56:418–422. doi:10.1016/S0090-4295(00)00672-5
11. Ge P, Gao F, Chen G. Predictive models for prostate cancer based on logistic regression and artificial neural network.
12. Stephan C, Cammann H, Semjonow A, et al. Multicenter evaluation of an artificial neural network to increase the prostate cancer detection rate and reduce unnecessary biopsies. Clin Chem. 2002;48:1279–1287. doi:10.1093/clinchem/48.8.1279
13. Humphrey PA. Histopathology of prostate cancer. Cold Spring Harb Perspect Med. 2017;7(10):a030411. doi:10.1101/cshperspect.a030411
14. Waliszewski P, Wagenlehner F, Gattenlöhner S, Weidner W. On the relationship between tumor structure and complexity of the spatial distribution of cancer cell nuclei: a fractal geometrical model of prostate carcinoma. Prostate. 2015;75:399–414. doi:10.1002/pros.22926
15. Bhele S, Ma Z, Mohanty S, et al. A machine learning tool to complement Gleason grading of prostate carcinoma. 2014.
16. Lawrentschuk N, Lockwood G, Davies P, et al. Predicting prostate biopsy outcome: artificial neural networks and polychotomous regression are equivalent models. Int Urol Nephrol. 2011;43:23–30. doi:10.1007/s11255-010-9750-7
17. Rodvold DM, McLeod DG, Brandt JM, Snow PB, Murphy GP. Introduction to artificial neural networks for physicians: taking the lid off the black box. Prostate. 2001;46:39–44. doi:10.1002/1097-0045(200101)46:1<39::AID-PROS1006>3.0.CO;2-M
18. Remzi M, Anagnostou T, Ravery V, et al. An artificial neural network to predict the outcome of repeat prostate biopsies. Urology. 2003;62:456–460. doi:10.1016/S0090-4295(03)00409-6
19. Moore CM, Petrides N, Emberton M. Can MRI replace serial biopsies in men on active surveillance for prostate cancer? Curr Opin Urol. 2014;24:280–287. doi:10.1097/MOU.0000000000000040
20. Sato C, Naganawa S, Nakamura T, et al. Differentiation of noncancerous tissue and cancer lesions by apparent diffusion coefficient values in transition and peripheral zones of the prostate. J Magn Reson Imaging. 2005;21:258–262. doi:10.1002/jmri.20251
21. Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A. 2015;112:E6265–6273. doi:10.1073/pnas.1505935112
22. Antonelli M, Johnston EW, Dikaios N, et al. Machine learning classifiers can predict Gleason pattern 4 prostate cancer with greater accuracy than experienced radiologists. Eur Radiol. 2019;29:4754–4764. doi:10.1007/s00330-019-06244-2
23. Toivonen J, Montoya Perez I, Movahedi P, et al. Radiomics and machine learning of multisequence multiparametric prostate MRI: towards improved non-invasive prostate cancer characterization. PLoS One. 2019;14:e0217702. doi:10.1371/journal.pone.0217702
24. Choyke PL, Loeb S. Active surveillance of prostate cancer. Oncology (Williston Park). 2017;31:67–70.
25. Jin W, Fei X, Wang X, Song Y, Chen F. Detection and prognosis of prostate cancer using blood-based biomarkers. Mediators Inflamm. 2020;2020:8730608. doi:10.1155/2020/8730608
26. McDonald AC, Vira M, Shen J, et al. Circulating microRNAs in plasma as potential biomarkers for the early detection of prostate cancer. Prostate. 2018;78:411–418.
27. Martignano F, Rossi L, Maugeri A, et al. A-based biomarkers for prostate cancer detection. Clin Chim Acta. 2017;473:96–105. doi:10.1016/j.cca.2017.08.009
28. Jakobsen NA, Hamdy FC, Bryant RJ. Novel biomarkers for the detection of prostate cancer. J Clin Urol. 2016;9:3–10. doi:10.1177/2051415816656121
29. Filella X, Foj L. Prostate cancer detection and prognosis from Prostate Specific Antigen (PSA) to exosomal biomarkers. Int J Mol Sci. 2016;17.
30. Reed AB, Parekh DJ. Biomarkers for prostate cancer detection. Expert Rev Anticancer Ther. 2010;10:103–114. doi:10.1586/era.09.168
31. Margreiter M, Stangelberger A, Valimberti E, Herwig R, Djavan B. Biomarkers for early prostate cancer detection. Minerva Urol Nefrol. 2008;60:51–60.
32. Parekh DJ, Ankerst DP, Troyer D, Srivastava S, Thompson IM. Biomarkers for prostate cancer detection. J Urol. 2007;178:2252–2259. doi:10.1016/j.juro.2007.08.055
33. Shariat SF, Karam JA, Roehrborn CG. Blood biomarkers for prostate cancer detection and prognosis. Future Oncol. 2007;3:449–461. doi:10.2217/14796694.3.4.449
34. Stephan C, Cammann H, Meyer HA, Lein M, Jung K. PSA and new biomarkers within multivariate models to improve early detection of prostate cancer. Cancer Lett. 2007;249:18–29. doi:10.1016/j.canlet.2006.12.031
35. Fruge AD, Smith KS, Bail JR, Rais-Bahrami S, Demark-Wahnefried W. Biomarkers associated with tumor Ki67 and Cathepsin L gene expression in prostate cancer patients participating in a presurgical weight loss trial. Front Oncol. 2020;10:544201. doi:10.3389/fonc.2020.544201
36. Green WJ, Ball G, Hulman G, et al. KI67 and DLX2 predict increased risk of metastasis formation in prostate cancer – a targeted molecular approach. Br J Cancer. 2016;115:236–242. doi:10.1038/bjc.2016.169
37. Kim Y, Jeon J, Mejia S, et al. Targeted proteomics identifies liquid-biopsy signatures for extracapsular prostate cancer. Nat Commun. 2016;7:11906.
38. Auffenberg GB, Ghani KR, Ramani S, et al. Urological Surgery Improvement, askMUSIC: leveraging a clinical registry to develop a new machine learning model to inform patients of prostate cancer treatments chosen by similar men. Eur Urol. 2019;75:901–907. doi:10.1016/j.eururo.2018.09.050
39. Alitto AR, Gatta R, Vanneste B, et al. PRODIGE: pRediction models in prOstate cancer for personalized meDIcine challenGE. Future Oncol. 2017;13:2171–2181. doi:10.2217/fon-2017-0142
40. Neeraj K, Ruchika V, Ashish A, et al. Convolutional neural networks for prostate cancer recurrence prediction. Proc SPIE. 2017.
41. Naguib RN, Robinson MC, Neal DE, Hamdy FC. Neural network analysis of combined conventional and experimental prognostic markers in prostate cancer: a pilot study. Br J Cancer. 1998;78:246–250. doi:10.1038/bjc.1998.472
42. Gustavsen G, Gullet L, Cole D, Lewine N, Bishoff JT. Economic burden of illness associated with localized prostate cancer in the United States. Future Oncol. 2020;16:4265–4277. doi:10.2217/fon-2019-0639
43. Liberati A, Altman D, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Journal of clinical epidemiology. 2009;62:(10).
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
