Back to Journals » International Journal of Nanomedicine » Volume 18
A Review of Labeling Approaches Used in Small Extracellular Vesicles Tracing and Imaging
Authors Bao C , Xiang H, Chen Q, Zhao Y, Gao Q, Huang F, Mao L
Received 6 April 2023
Accepted for publication 26 July 2023
Published 11 August 2023 Volume 2023:18 Pages 4567—4588
DOI https://doi.org/10.2147/IJN.S416131
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mian Wang
Chenxuan Bao,1 Huayuan Xiang,1 Qiaoqiao Chen,1,2 Yuxue Zhao,1,2 Qianqian Gao,1 Feng Huang,1 Lingxiang Mao1,2
1Department of Laboratory Medicine, Affiliated Kunshan Hospital of Jiangsu University, Kunshan, Jiangsu, People’s Republic of China; 2Department of Laboratory Medicine, the Affiliated People’s Hospital, Jiangsu University, Zhenjiang, Jiangsu, People’s Republic of China
Correspondence: Lingxiang Mao, Department of Laboratory Medicine, Affiliated Kunshan Hospital of Jiangsu University, Kunshan, Jiangsu, People’s Republic of China, Tel +86 15358599331, Email [email protected]
Abstract: Small extracellular vesicles (sEVs), a subset of extracellular vesicles (EVs) originating from the endosomal compartment, are a kind of lipid bilayer vesicles released by almost all types of cells, serving as natural carriers of nucleic acids, proteins, and lipids for intercellular communication and transfer of bioactive molecules. The current findings suggest their vital role in physiological and pathological processes. Various sEVs labeling techniques have been developed for the more advanced study of the function, mode of action, bio-distribution, and related information of sEVs. In this review, we summarize the existing and emerging sEVs labeling techniques, including fluorescent labeling, radioisotope labeling, nanoparticle labeling, chemical contrast agents labeling, and label-free technique. These approaches will pave the way for an in-depth study of sEVs. We present a systematic and comprehensive review of the principles, advantages, disadvantages, and applications of these techniques, to help promote applications of these labeling approaches in future research on sEVs.
Keywords: small extracellular vesicles, label, image, tracking, in vivo imaging
Introduction
Extracellular vesicles (EVs) are a collective term for particles wrapped by lipid bilayers naturally released from cells that cannot self-replicate. It includes several subtypes, with microvesicles, exosomes, and apoptotic vesicles being major subtypes.1 Exosomes, also known as intraluminal vesicles (ILVs), are small extracellular vesicles (sEVs) formed via the endosomal pathway and range from 30 to 200 nm in diameter. They contain a series of membrane-associated high-order oligomeric protein complexes that display significant molecular heterogeneity.2 Although sEVs have been discovered for over 80 years, they were initially considered only as metabolic waste products of cells until 2007, when Valad et al3 discovered that mRNA and microRNA could be transported to other receptor cells via sEVs and perform corresponding physiological functions. Since then, researchers have started to study sEVs in depth, and their biogenesis and physiological functions have started to be discovered continuously.
sEVs carrying specific proteins, lipids, nucleic acids, and glycoconjugates are part of an intercellular signaling network in multicellular tissues. They mediate multiple activities such as signaling and molecular transmission to other cells.2,4 sEVs can be secreted by almost all types of cells and have been identified in body fluids such as plasma, urine, semen, saliva, bronchial fluid, cerebrospinal fluid, breast milk, serum, amniotic fluid, synovial fluid, tear fluid, lymphatic fluid, bile, and gastric acid.5 This pathway of sEVs transport plays an essential role in many aspects of human health and disease, including development, immunity, tissue homeostasis, cancer, and neurodegenerative diseases. In addition, viruses select the sEVs biogenesis pathway to assemble infectious particles and establish host compatibility.2 Because of their known physiological and pathological functions, their clinical application as biomarkers, therapeutic agents, and therapeutic molecular carriers are considered widely promising and have yielded notable results.4
The formation, release, recycling, uptake, and content delivery of sEVs have been critical aspects of sEVs research, and the translation and application of research results are still in their infancy. The study of these problems requires the help of adequate sEVs labeling and tracer imaging technique, and the appropriate labeling technique can help researchers to investigate the biological properties of sEVs in depth. Therefore, effective in vivo and in vitro sEVs labeling techniques are essential. This paper provides a review of sEVs labeling techniques.
Membrane Fluorescent Dye Labeling Technique
Lipophilic Fluorescent Dyes
Carbonaceous Dyes
Carbonaceous dyes are a class of lipophilic fluorescent dyes that can be used to stain cell membranes and other lipid-soluble biological structures, including membranes of sEVs. Their fluorescence intensity is greatly enhanced when bound to lipid membranes, and these dyes have high quenching constants and long excited state lifetimes. Once the lipid bilayer is stained, these dyes diffuse throughout the whole membrane and can be applied to the entire membrane structure of sEVs at optimal concentrations. Common carbocyanine dyes include DilC18(5) (DiD) (λex/λem=646/663 nm), DilC18(3) (DiI) (λex/λem=551/569 nm), DiOC18(3) (DiO) (λex/λem=483/501 nm), and DilC18(7) (DiR) (λex/λem=754/778 nm). DiI and DiO are widely used for in vitro tracing of sEVs due to their very weak fluorescent signal before entering the membrane and the intense signal after entering the membrane structure. Yu et al6 used DiI to identify the uptake of sEVs by bacteria in their study of the effect of different types of sEVs on bacterial growth. Murdica et al7 used DiO-labeled endometrial cells and found that their derived sEVs enhanced the ability of sperm to capacitate and acrosome react. And discovering an emerging way for female genitourinary tract cells to interact with sperm. On top of DiI and DiO, DID and DIR have a lower background fluorescence, while their staining is more stable, and almost no staining transfer between sEVs occurs. More importantly, its longer Near Infrared (NIR) wavelength gives it better results in vivo experiments due to the significant reduction in autofluorescence of animals at higher wavelengths.8 Based on this property, this dye has been used in both in vivo and in vitro studies of sEVs.8–10 Santelices et al9 detailed the labeling procedure of sEVs obtained from in vitro culture using DIR. Wen et al10 used DID to achieve in vivo imaging of MSC-EVs in mice and verified the accumulation of MSC-EVs at radiation-damaged sites in mice.
FM Styryl Dyes
FM dyes are a type of lipophilic styrene-based compound with bipolarity. Hydrophilic grouping effectively avoids the occurrence of membrane penetration staining and intercellular cross-staining. Thus, they are widely used in experiments related to membrane labeling and vesicle formation. FM dyes include FM 1–43 (N-(3-Triethylammoniumpropyl)-4-(4-(dibutylamino) styryl)pyridinium dibromide) (λex/λem=510/626 nm) and FM 4–64(N-(3-Triethylammoniumpropyl)-4-(6-(4-(Diethylamino) Phenyl) Hexatrienyl) Pyridinium Dibromide) (λex/λem=558/734 nm). Wolf et al11 used FM 4–64-labeled bovine milk sEVs in human colon cancer cells Caco-2 and rat small intestinal cells IEC-6 to elucidate the mechanism by which intestinal transport of bovine milk sEVs is mediated by endocytosis.
PKH Dyes
PKH dyes are another class of lipophilic dyes commonly used for sEVs tracing, invented by Paul Karl Horan12 and hence the name. Their two long lipophilic tails can be inserted into the lipid bilayer structure, while the fluorescent groups emit fluorescence outside the lipid membrane. Commonly used PKH dyes include PKH26 (λex/λem=551/567 nm) and PKH67 (λex/λem=490/502 nm). PKH dyes can also be used in in vivo and in vitro studies of sEVs.13–15 Reclusa et al13 compared PKH26 with PKH67 for labeling imaging during phagocytosis of sEVs in lung cancer cells and observed that better image intensity and contrast were achieved using PKH67 for labeling sEVs. Unfortunately, the underlying causes for this phenomenon have not been extensively investigated in the corresponding article. Pollalis et al,15 on the other hand, exploited the in vivo imaging capability of PKH26 and found the targeting of murine retinal sEVs delivery capability. Although PKH series dyes are now widely used for sEVs labeling, there is a growing consensus that the PKH series may not be suitable, Dehghani et al16 found that using PKH to label sEVs severely affects the particle size of sEVs. Also, the aggregation effect of PKH dyes is very significant. It can be taken up by cells, leading to false positives, which is detrimental to the analysis of the results.17 The use of PKH dyes is worth considering during future sEVs studies.
Nobel Lipophilic Fluorescent Dye
To overcome the typical defects of traditional dyes like PKH, Shimomura et al18 reported a class of lipophilic fluorescent dyes called Mem (λex/λem = 494–652 nm/512-672 nm). Mem synthesis contains glutamic acids to avoid probe penetrating and aggregation. They were verified by experimenters to show that they did not affect the size of sEVs and did not form aggregation. The CellMaskTM (λex/λem=522–649 nm/535-666 nm) series of dyes is a commercially available lipophilic dye whose most remarkable feature is slow internalization and can be used for coloration after cell fixation. However, the detergent will destroy the structure of CellMaskTM. Therefore, they cannot be used with probes requiring permeabilization. El-Andaloussi et al19 employed CellMaskTM for tracing sEVs to monitor their uptake in an experimental method, they introduced for delivering siRNA in vitro using sEVs. In 2019, Collot et al20 invented a series of six fluorescent dyes called MemBrightTM (Figure 1A) with a long wavelength range (λex/λem=499–689 nm/506-713 nm, Figure 1B). These new membrane probes are based on zwitterionic anchors and six dialkyne cyanine fluorophores. Due to its amphiphilicity, it forms soluble aggregations in aqueous media and quenches spontaneously until it dissociates in contact with lipids and emits fluorescence (Figure 1C). Its working concentration is exceptionally low, and nanomolar concentration dye can stain plasma membrane structures exactly. Compared to conventional lipophilic dyes, it has the advantages of simpler handling, greater specificity, lower working concentration, and no cytotoxicity. It could be used with live/fixed cells for immunostaining and is compatible with different fluorescence imaging techniques such as long-term, two-photon, tissue, and super-resolution imaging. Subsequently, Hyenne et al21 verified the feasibility of tracking circulating tumor EVs in zebrafish embryos in vivo (Figure 1D). The following year, Boyer et al22 also applied MemBrightTM dye for labeling and tracing sEVs in a study that confirmed that endothelium-derived sEVs regulate the phenotype of vascular smooth muscle cells.
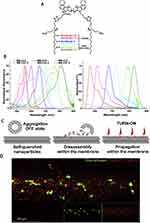 |
Figure 1 (A) Structure of the MemBrightTM markers. (B) Absorption and emission spectra of MemBrightTM probes (200 nM) in the absence (dashed lines) or presence of DOPC vesicles. (C) Turn-on mechanism of the MemBrightTM probes. (A)-(C) Reprinted from Cell Chem Biol, 26(4), Collot M, Ashokkumar P, Anton H, et al. MemBright: a family of fluorescent membrane probes for advanced cellular imaging and neuroscience. 600–614 e7, Copyright (2019), with permission from Elsevier.20 (D) Confocal images of MemBright-Cy3 labeled Zmel1 EVs 3 hpi in Tg (mpeg1: GFP) (macrophage specific expression). Reprinted from Dev Cell, 48(4), Hyenne V, Ghoroghi S, Collot M, et al. Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. 554–572 e7, Copyright (2019), with permission from Elsevier.21 |
Membrane Analog
The effective staining labeling of sEVs membranes can also be formed by fluorescently labeled analogs of lipid membrane components. Verderio et al23 reported a technique for successful labeling of NBD-C6-HPC, a phosphorylcholine analog, onto bone marrow microvesicular membranes. Similarly, Hu et al24 reported a cholesterol analog labeling dye called 3-NBD-cholesterol. It is verified in their experiments that they could form effective labels for sEVs by co-culturing labeled platelet sEVs with phagocytes and HEK293 cells and can be detected by fluorescence microscopy and flow cytometry. Like the carbocyanine dyes, NBD (λex/λem=463/536 nm) also forms effective fluorescent labeling only after entering the membrane structure, avoiding high background, misleading labeling, and other phenomena.
Fluorescent Lectins
Lectins are a class of naturally occurring proteins with hemagglutinating activity and sugar specificity. Because of its ability to bind glycoconjugates and glycoproteins in cell membrane structures, it is widely used as a tool for membrane and cell research.25 Combining lectins with fluorescent moieties for labeling cell membranes is widely used. Wheat germ agglutinin (WGA), which binds to N-acetylglucosamine and N-acetylneuraminic acid (sialic acid), is the most widely used. Lennon et al26 used WGA-647 (λex/λem=650/670 nm) to label pancreatic cancer EVs captured by Ab cetuximab because of their overexpression of epidermal growth factor receptor 1 (EGFR) and successfully applied quantitative single-molecule localization microscopy (qSMLM) to image them.
Self-Quenched Probe for Surveying Membrane Fusion
Octadecyl Rhodamine B Chloride (R18) (λex/λem=560/590 nm) is a lipophilic probe that binds to membranes with its fluorescent moiety located in the aqueous phase layer and the alkyl tail extending into the inner lipid layer. R18 fluorescence is quenched at high concentrations and is gradually released once the concentration is diluted. This feature makes it a probe for membrane fusion studies. Tian et al27 used this feature to distinguish between two different modes of sEVs uptake: endocytosis and membrane fusion.
Traditional lipid membrane dyes have been widely used as tracing markers in sEVs research due to their early application, relatively simple experimental procedures, low cost, strong stability, and long half-life. However, as their practical applications have progressed, more and more issues have emerged. For example, (i) current methods for isolating sEVs struggle to remove lipid contaminants, inevitably leading to nonspecific staining. (ii) Lipophilic dyes, due to their molecular characteristics, easily form aggregates that can affect the interpretation of experimental results. This situation is particularly severe when using samples obtained through ultracentrifugation. (iii) Lipid membrane dyes used for labeling sEVs may impact membrane fusion with cells, the physiological properties of the membrane, the texture of the membrane, and the size of sEVs. (iv) Due to dye transfer and their much longer half-life compared to the lifespan of sEVs, there can be an impact on the study of sEVs outcomes within cells. Some researchers recommend avoiding the use of membrane dyes for labeling sEVs of plasma samples obtained through ultracentrifugation. Additionally, sEVs labeling experiments should involve thorough washing to remove unbound dye and require careful experimental design with appropriate controls to correctly interpret the results. Considering the poor performance of PKH dyes in experiments, their use for sEVs labeling needs to be carefully considered. Overcoming the limitations of traditional dyes, novel dyes with self-quenching, small molecular size, and high affinity should be of interest and consideration for researchers.16,17,28–33
Bio-Conjugation Labeling Technique
Amine-Reactive Dyes
Amine-reactive dyes are a class of tracer dyes that are labeled by amide reactions. Carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) is the most commonly used amine-reactive dye for sEVs tracing. CFDA-SE (λex/λem=500 nm/520 nm) CFSE can easily penetrate cell membranes, covalently bind to intracellular proteins in living cells, and release green fluorescence after hydrolysis. The succinimidyl ester reacts with the intracellular amino group to form a stable fluorescent conjugate that can be fixed with an aldehyde fixator. Due to the labeling located inside the cells and the sEVs, it does not affect the size and function of sEVs, and the labeling is effective for several weeks.16,34 Radnaa et al35 labeled sEVs containing high mobility group box 1 protein (HMGB1) with CFDA-SE to study the mechanism of this sEVs molecule. Notably, the inherent properties of carboxyfluorescein, whose fluorescence intensity is too weak in an acidic environment, prevent its application for monitoring the exocytosis of sEVs in the endosomal pathway.36 Given that the defect of CFDA-SE, Zhou et al37 developed a novel amine-reactive dye called ExoTracker (λex/λem = 550 nm/670 nm), which is conjugated to the sEVs membrane by amide reaction and undergoes structural changes in this process to form a fluorescent label. Its pH characteristics are opposite to those of CFDA-SE. It has a better fluorescent ability in acidic environments, thus complementing CFDA-SE. Besides, Tian et al38 used an amine-reactive dye called TAMRA-NHS (carboxytetramethylrhodamine succinimidyl ester) (λex/λem=553 nm/575 nm), whose NHS binds to r membrane proteins of sEVs to achieve a labeling tracer effect when studying the fate of protein components of sEVs being uptaken by recipient cells.
Thiol-Reactive Dyes
Thiol-reactive dyes are a class of tracer dyes that label sEVs by combining fluorescent groups with maleimide. This organic compound can combine with thiol groups in tetraspanins on the sEVs.39 Roberts-Dalton et al40 used Alexa488 (λex/λem=494 nm/517 nm) combined with maleimide to form a novel fluorescent dye to label sEVs and then uptake by Hela cells. They found that sEVs are uptaken by clathrin-independent endocytosis and ultimately delivered to the lysosome. Labelling of EVs in this way did not influence their size and had no effect on their abilities.
Azide-Alkyne-Cycloaddition Labeling Technique
The azide-alkyne-cycloaddition reaction is a click chemical reaction between the azide and the strained alkyne ring, which is catalyzed by the copper to form a triazole bond. A copper-free reaction method was also developed for biological research.41 There are three primary forms of this labeling method: i) Direct labeling of isolated sEVs with chemicals containing azides and alkynes. Xu et al42 successfully labeled PANC-1 Exo through the reaction between the N-hydroxysuccinimidyl (NHS) group of dibenzobicyclooctyne (DBCO) and amine groups of lysine residues on sEVs surface proteins (Figure 2A). ii) Supplementing sEVs-producing cells with azide- or alkyne-containing amino acids to produce azide- or alkyne-containing proteins by the cells’ own biosynthesis and adding the corresponding compound with the labeling group to label the cells and their sEVs. iii) Supplementing sEVs-producing cells with azide- or alkyne-containing amino acid glycans/proteoglycans to generate membrane surface glycans containing azide or alkyne. Then add the corresponding compound with the labeling group to label the cells and their sEVs. Wang et al43 used a residue-specific protein labeling strategy. They co-cultured sEVs-producing cells with L-azidohomoalanine (AHA), an azide-bearing amino acid analog of methionine and tetra-acetylated N-azidoacetyl-D-mannosamine (ManNAz), an azidosugar that can be metabolized into sialic acid. Unnatural azides were introduced into sEVs through protein and sugar metabolism. Then, the addition of DBCO with reporting groups to the culture system achieved valid labeling of producing sEVs (Figure 2B). Because of the relevance of this labeling technique for glycan metabolism at the cell membrane surface, this technique has also been used for glycan imaging and sEVs surface glycan analysis.44,45 Based on the high flexibility of click chemistry, different labeling types can be easily achieved by substituting reporter moieties and enabling further modification of the composition and functionality of sEVs. Providing novel, powerful tools to study the roles of sEVs in biology and expand the biomedical potential of sEVs.
 |
Figure 2 (A) Scheme of the fluorescence labelling method of sEVs surface using copper-free click chemistry with AlexaFlour®488 (AF488)-azide. Reprinted from Xu L, Faruqu FN, Liam-Or R, et al. Design of experiment (DoE)-driven and uptake studies of exosomes for pancreatic cancer delivery enabled by copper-free click chemistry-based labelling. J Extracell Vesicles. 2020;9(1):1779458. © 2020 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group on behalf of The International Society for Extracellular Vesicles. Creative Commons.42 (B)Combination the metabolic labeling of newly synthesized proteins or glycan/glycoproteins of exosome-secreting cells with chemically active azide groups and biorthogonal click conjugation to modify and functionalize sEVs. Reprinted from Wang M, Altinoglu S, Takeda YS, Xu Q. Integrating Protein Engineering and Bioorthogonal Click Conjugation for Extracellular Vesicle Modulation and Intracellular Delivery. PLoS One. 2015;10(11):e0141860. Creative Commons.43 |
Non-Covalent Labeling Technique
Immunolabeling
Immunolabeling is also a commonly used labeling technique. The main principle is to label the sEVs target protein with the corresponding primary antibody and then bind the primary antibody with the secondary antibody coupled with a fluorescent group. Then the sEVs are valid labeled. Tetraspanins are specific sEVs membrane proteins that are the most commonly used primary antibody-binding proteins. Chen et al46 labeled sEVs secreted by breast cancer cells using secondary antibodies coupled to Alexa Fluor 647 with primary antibodies recognizing CD63 proteins (Figure 3A). Besides Tetraspanins, sEVs-specific proteins such as TSG101 and HSP7047 or target sEVs-specific proteins like HER-246 can also be labeled as primary antibody-binding proteins. Mondal etal47 took glioblastoma (GB) cell-line KW10 as an example to describe immunofluorescence labeling techniques for sEVs membrane-specific proteins CD63, TSG101 and calnexin in detail. Based on antigen-antibody reactions, immunolabeling exhibits strong specificity, allowing effective discrimination of different sources of sEVs or different subpopulations of sEVs as needed. However, this method also has some drawbacks. Due to the small diameter of sEVs, the binding of a large number of antibodies and labeled proteins on the surface of sEVs will increase the size of sEVs and obscure other functional receptors on their surface, thus seriously affecting their physicochemical properties and biological functions.
 |
Figure 3 (A) Schematic illustration of indirect IF labeling of CD63 and sEVs membrane stained with CM-Dil. Reprinted with permission from Chen C, Zong S, Wang Z, et al. Imaging and Intracellular Tracking of Cancer-Derived Exosomes Using Single-Molecule Localization-Based Super-Resolution Microscope. ACS Appl Mater Interfaces. 2016;8(39):25825–25833. Copyright © 2016 American Chemical Society.46 (B) Schematic of aptamer-based DNA nanoassemblies on the surfaces of sEVs. Reprinted with permission from Wan S, Zhang L, Wang S, et al. Molecular Recognition-Based DNA Nanoassemblies on the Surfaces of Nanosized Exosomes. J Am Chem Soc. 2017;139(15):5289–5292. Copyright © 2017 American Chemical Society.48 (C) Schematic illustration of screening for sEVs anchor peptides and functionalization on sEVs. Reprinted from Gao X, Ran N, Dong X, et al. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci Transl Med. 2018;10:444. Creative Commons.49 |
Labeling by Aptamers
The aptamer is a short single-stranded nucleic acid molecule typically generated by the systematic evolution of ligands by exponential enrichment (SELEX). The desired aptamers can then be selected in the synthetic library. The aptamer can specifically bind to target molecules in a similar manner to antigen–antibody binding.50 Wan et al48 reported an aptamer-based nanolabeling technique on the surface of sEVs, which selected the HepG2 cell-specific binding aptamer LZH8, and achieved specific labeling of HepG2 sEVs by hybridization chain reaction with two nucleic acid monomers to form a switch structure and then added fluorescein isothiocyanate (FITC) to achieve fluorescent labeling of HepG2 sEVs (Figure 3B). The authors claimed that this technique could reach different types of labeling by changing the labeling group. Zhu et al44 reported a dual labeling method for aptamer and orthogonal chemical labeling. They achieved dual labeling of exoPD-L1 on sEVs surfaces, respectively, through orthogonal chemistry labeling by glycan metabolism with alkyne DBCO-Cy5 and PD-L1-specific aptamer with Cy3. They first demonstrated that glycosylation of exoPD-L1 is the structural basis for exoPD-L1/PD-1 interaction and inhibition of CD8+T cell proliferation. It provides a theoretical basis for immunotherapy. Compared with antigen–antibody reactions, aptamer molecules are smaller, have higher biocompatibility, and can effectively avoid the impact of labeling on sEVs physicochemical properties and biological functions.51
Labeling by Anchoring CP05 Peptide
CP05 peptide has a strong affinity for the extracellular second ring structure of CD63, an sEVs-specific protein. Hence, it can be used as an effective linkage between the marker molecule and sEVs. Gao et al49 verified the in vitro tracer ability of CP05 by co-incubating CP05 labeled by Radamine with sEVs. They bound AF680-labeled CP05 to various sEVs, demonstrating its in vivo tracer and nonspecifically-binding sEVs properties. They also labeled Rhodamine-labeled M12-CP05, a muscle-targeting peptide, and FITC-labeled NP41-CP05, a nerve-targeting peptide with sEVs, verifying CP05’s dual labeling capability and no interaction between the different CP05 markers (Figure 3C). As a short peptide, CP05 has minimal impact on the functionality and size of sEVs. Additionally, its production difficulty is relatively lower compared to monoclonal antibodies, and the subsequent conjugation of molecules to CP05 is simpler.
Labeling by Electrostatic Interactions
The sEVs surface is considered to be negatively charged overall due to its richness in proteoglycans, sialic acid, and lipids. This allows us to label sEVs by making the target molecule positively charged.52–54 DPA-SCP is a positively charged aggregation-induced emission luminogens (AIEgens) that can be directly adsorbed to the sEVs membrane surface via electrostatic interaction. Using this technique, Cao et al55,56 injected DPA-SCP-labeled MSC-EVs into liver-injured or kidney-injured mice and successively observed specific aggregation of MSC-EVs in the injured organs, verifying the in vivo and in vitro labeling ability of AIEgens. The experimental results demonstrate that, apart from slightly altering the membrane potential of sEVs, no significant impact on the size and functionality of sEVs was observed. Additionally, compared to traditional labeling methods such as PKH26 and DiO, it exhibited superior imaging effects.
Labeling sEVs via Cell-Engineering Technique
Biomarkers such as tetraspanins, heat shock proteins (HSPs), lactadherins, and donor cell-specific proteins like viral envelope protein VSVG57 are present on the sEVs membrane surface. By integrating the coding sequences of fluorescent proteins or luciferases with those of these proteins through genetic engineering techniques, the corresponding fusion proteins can be constructed to label and trace sEVs under specific conditions.
Labeling by Fluorescent Protein
The fluorescent protein is a commonly used reporter protein that emits a corresponding fluorescent signal in response to excitation light of a specific wavelength. We can trace the tagged sEVs by constructing a fusion protein from reported fluorescent protein fusions to sEVs proteins. Currently, a fusion protein that binds CD63 to a reporter protein is widely used. Men et al58 constructed a mouse model expressing CD63-GFP fusion protein and determined the localization of neuron-secreted sEVs in astrocytes. Similarly, Li et al59 used a CD63-iRFP682 fusion protein to establish a breast cancer cell line that stably expresses sEVs containing this protein. With the exception of the traditional form of fusion proteins, scholars have developed a new type of fusion protein. Fluorescent proteins such as EGFP, tdTomato, and mNeonGreen60,61 are bound to the palmitoyl group. Then, with the catalyzation by the intrinsic palmitoyl acyltransferase (PAT)62 of somatic cells, the palmitoyl group is bound to thiol groups on the surface of the sEVs membrane. Then the sEVs are bound by the fluorescent protein.
Labeling by Bioluminescence
The bioluminescent labeling technique is also called the luciferase labeling technique. Its principle is similar to fluorescent proteins. However, unlike fluorescence imaging, which requires excitation light, luciferases can emit light spontaneously when combined with specific substrates. Common luciferase genes include Fluc, Gluc, Rluc, and Nluc.63–66 For example, Takahashi et al64 established a reporter model of a gLuc-LA fusion protein, expressed in mouse melanoma B16BL6 cells. Collecting the sEVs containing fusion protein and injecting them into mice through intravenous injection. They found that sEVs were cleared by macrophage uptake in the liver, spleen, and lung. In their study, Gupta et al28 compared the labeling effects of five luciferases, NanoLuc, ThermoLuc, Fluc, Super Rluc8, and CBRLuc. The results show higher luminescence efficiency and more stable labeling of NanoLuc, while ThermoLuc has better performance in in vivo experiments due to its NIR emission wavelength. Both of them have higher labeling efficiency for sEVs. Therefore, the authors recommend using NanoLuc for sEVs labeling in vitro and ThermoLuc for tracing in vivo.
In order to avoid the limitations of using a single fusion protein, two or even multiple-reporter genes could be used simultaneously to label sEVs-producing cells to homogenize the monitoring levels or to monitor multiple components simultaneously.67,68 Notably, when multiple fusion proteins label the same sEVs-producing cells, different fusion proteins have different loading efficiencies and specificities into the secreted sEVs.69 Genetically, engineered labeling technique has high specificity, high flexibility, low impact on cells, and high stability compared to traditional membrane dye labeling technique. Although the sEVs of clinical samples are difficult to trace from the source by this technique, its extremely flexible and stable labeling modalities make it promising for the study of sEVs biogenesis, in vivo transport, cellular uptake, and eventual fate.
Radiolabeling
Radioisotope labeling is a classical laboratory labeling technique that has been exploited and applied in sEVs tracing research. Compared with traditional optical labeling techniques, radioactive labeling has high sensitivity and stable imaging performance in tissue imaging, in vivo imaging, and even clinical imaging. The commonly used isotopes for sEVs labeling include 131I, 125I, 124I, 111In, 99mTc, 89Zr, 68Ga, 64Cu, and so on. These signals can be detected by single photon emission computed tomography (SPECT) or positron emission tomography (PET) noninvasively. The isolated organs can also be imaged by Computed Tomography (CT) or Magnetic Resonance (MR).70
Iodine radioisotopes are an extensively used class of isotopes for labeling. Rashid et al70 showed the distribution of radioisotope 131I-labeled tumor sEVs (TDEs) in vivo from different cellular origins through SPECT/CT. Morishita et al71 combined isotopic labeling with genetic engineering techniques to form 125I labeled sEVs via a streptavidin-lactadherin fusion protein and 125I labeled biotin and found that the liver predominantly cleared B16BL16-derived sEVs. Royo et al72 directly labeled sEVs with [124I]NaI and monitored them by PET. They demonstrated that sEVs accumulated rapidly in the liver and were present in multiple organs, including the brain.
Despite iodine, metal isotopes are also widely used. Faruqu et al73 reported two sEVs labeling techniques based on 111In. One is an endo-labeling technique with tropolone-mediated isotope penetration through the membrane into the sEVs lumen; the other is a membrane labeling technique based on amine-responsive DTPA-anhydride bidirectional binding of sEVs membranes and isotopes. In comparison, the membrane label has better effect and flexibility. 99mTc is a more used isotope in the field of sEVs labeling. Gonzalez et al74 reported a 99mTc-based ionic salt (99mTcCl4) labeling approach. Compared to the previous complex compound labeling like 99mTc-HMPAO,75 (99m)Tc-tricarbonyl,76 and [99mTc(CO)3(H2O)3]+,77 ionic salt labeling has a more simpler operation and lower cost, while also ensuring labeling efficiency. Recently, 89Zr-based sEVs isotope labeling tracer methods have also been reported, including directly labeling sEVs membrane with zirconium-89 (89Zr) via amine reactions78 and endo-labeling with [89Zr]Zr(oxinate)4.79 Besides, Jung et al80 successfully visualized the biodistribution of sEVs in the lymphatic or bloodstream pathways using68 Ga or 64Cu-labeled sEVs and demonstrated the higher sensitivity of isotopic labeling, compared with Cy7 fluorescent labeling. Warashina et al81 also labeled sEVs with 64Cu via a cross-bridged macrocyclic chelator (CB-TE1A1P) for pharmacokinetic study and determined the contribution of macrophages in the uptake of sEVs by the liver and spleen. In addition, Horgan et al82 reported an sEVs molecular imaging technique in vitro based on Raman metabolic labeling. Deuterium-labeled sEVs are formed by the cell’s own metabolism and used for subsequent tracing of cellular uptake for internalization and other studies.
Although radioisotopes labeling in deep tissues has advantages that are difficult to match with conventional labeling methods, its application on sEVs labeling is relatively few. On the one hand, the use of radioactive substances requires large precision instruments and higher operational capability; on the other hand, the use of radioactive substances is under strict surveillance, especially for clinical applications. In addition, when utilizing radiolabeling of sEVs with radioisotope, appropriate chelators or alternative methods should be selected. Direct labeling of sEVs with radioisotopes exhibits drawbacks like detachment after in vivo injection or potential impact of radioisotopes and labeling process on the functionality of sEVs. Suitable labeling of media or treatments is a critical issue in the process of radiolabeling. It is also a primary focus for subsequent research endeavors.
Nanoparticles Labeling
Metal Nanoparticles Labeling
Metal nanoparticles are excellent and widely used contrast agents for sEVs imaging. Gold is an excellent contrast metal for CT, MRI, and Photoacoustic Imaging (PAI). Popovtzer’s team83–85 developed glucose-encapsulated gold nanoparticles (Glu-GNPs) that could be directly transported into sEVs for CT imaging. They demonstrated that MSC-EVs could cross the blood–brain barrier and accumulate in various types of neuropathic regions through experiments. The results provide new technical support for the diagnosis and treatment of CNS diseases, on the one hand, and fill the gap in CT imaging localization of CNS diseases, on the other hand. Similarly, Lara et al86 established another method of CT imaging for sEVs using nanogold. They exploited the overexpression of folate receptors on the surface of B16F10 cells and conjugated nanogold with folate to generate AuNP-PEG-FA, which was endocytosed by cells through the folate receptor pathway and then entered their secreted sEVs through biosynthesis. Using this method, they observed that sEVs preferentially accumulated in small lung metastatic tumors and were precisely distributed along the tumor tissue. In addition, gold can also be used as a PAI photothermal contrast agent, combining both PAI contrast and photothermal therapy (PTT).87 Zhu et al88 co-incubated tumor cells with gold nanostars to produce a large number of tumor cell-derived stellate plasmonic sEVs (TDSP-Exos) that could be specifically taken up by tumor cells thereby PAI the tumor tissue and kill the tumor cells by PTT.
Superparamagnetic iron oxide nanoparticles (SPIONs), another widely used class of metallic particles, are magnetic particles ranging in size from 5 ~ 200 nm and are mainly used for MRI. Their superparamagnetic character is their most distinctive feature, allowing the movement of SPIONs to designated areas of the body to be controlled by an applied magnetics field.89 Their low toxicity, high sensitivity, and targeted delivery characteristics achieve better imaging results in the lesion area only at a low dose.90 First, due to its high biocompatibility, SPIONs can be attached to sEVs by various means such as electroporation, natural cellular uptake, or Bio-Conjugation and carry substances such as other labeling molecules or therapeutic drugs to meet experimental or therapeutic requirements.91 Busato et al92,93 labeled 100% of the cells using ultra-small superparamagnetic iron oxide nanoparticles (USPIO, 4–6 nm) and traced their secreted sEVs that retained USPIO labeling and physiological characteristics by MRI. Jia et al94 loaded SPIONs and curcumin into sEVs conjugated with neuromyelin-1 targeting peptide, which allows simultaneous imaging and therapy of glioma in vitro and in vivo. Zhang95 loaded sEVs conjugated with rabies virus glycoproteins with SPIONs to synthesize MRI contrast agent with targeting ability to nerve cells and successfully imaged them in vivo.
Some other metals also have the potential as contrast agents, such as manganese96,97 and gadolinium.98 Abello et al98 used lipid gadolinium to label sEVs for MRI and found a sustained accumulation of HUC-MSC sEVs in tumors. However, their application is limited due to the possible biotoxic effects of such metals.99
Quantum Dots Labeling
Quantum dots (QDs) are nanoscale semiconductors that emit light at a specific frequency by applying a particular electric field or optical pressure to this nanosemiconductor material. Compared to conventional organic dyes, they have long-term stability, efficient labeling, and “multiplexing.”100 Its synthetic customizability and broad excitation spectrum give it an extensive and flexible applicational prospect. Moreover, it has been used for imaging of sEVs in vivo and in vitro for decades.101 How to label QDs onto target sEVs is the key to QDs labeling techniques, and the related methods have been described previously. For example, sEVs can be labeled by QDs using antigen-antibody or aptamer techniques.102–105 Alternatively, QDs can be conjugated to sEVs using click chemistry106 or internalized into sEVs directly using the cells’ own uptake and internalization.107
The synthetic feature of Nanoparticles (NPs) gives great flexibility to this technique. Some scholars have used specifically synthesized NPs to implement multimodal imaging techniques. For example, Zhao et al107 synthesized ultra-small Mn magnetically functionalized Ag2Se quantum dots (Ag2Se@Mn QDs) that can be directly loaded into microvesicles (MVs) by electroporation. The labeled MVs can be traced by NIR and MRI. Bose et al108 synthesized similar NPs named GION for labeling TEVs for MRI and PTT. Shaikh et al109 performed CT, MRI, and fluorescence imaging (FLI) multimodal imaging of tumor cells and their derived sEVs using in situ generated iridium iron oxide nanoclusters (NCs). Cao et al110 synthesized vanadium carbide quantum dots (V2C QDs) loaded in engineered sEVs for FLI, PAI, MRI multimodal imaging, and tumor-targeted PTT. Tayyaba et al111 performed CT, MRI, and FLI multimodal imaging of sEVs from Ag-Fe304 NCs biosynthesized in situ from HepG2 cells.
Chemical Contrast Agent Labeling
In addition to metal NPs, some organic compounds or biomolecular substances also function as contrast agents. For example, chlorine6 (Ce6) is a photosensitizer that can be used for PAI. Although it is also a commonly used medicine for PTT, its clinical applications are limited by its low plasma concentration and nonspecific toxicity.112 Jiang et al113 loaded Ce6 into tumor-derived sEVs to form Ce6-R-Exo that could be targeted accumulated in tumor cells for PAI and PTT. Ferritin heavy chain (FTH1) is an MRI reporter protein.114 Liu et al115,116 achieved in vivo and in vitro MRI of target cell-derived sEVs by lentiviral transfection to make the target cells express FTH1.
Label-Free Visualization
The label-free technique, ie, visualizing sEVs without labeling, is usually based on the intrinsic properties of sEVs. The use of label-free technology avoids the influence of sEVs and allows them to be assayed in a state closer to normal physiology, while also reducing the experimental time. Currently, label-free techniques are mainly used for imaging, size analysis, and sorting of sEVs, such as TEM, NTA, DLS, FCM, AFM, and so on.117 And have been explored for liquid detection, characterization, and biochemical differences in content. These include Raman spectroscopy, surface enhanced raman spectroscopy (SERS), surface plasmon resonance spectroscopy (SPR), and infrared spectroscopy (FTIR).117,118 However, only some relevant research results in monitoring sEVs and imaging sEVs distribution exist. In this field, Boppart et al's119–122 research team achieved certain achievements in sEVs monitor imaging using multi-harmonic generation (MHG) and multiphoton autofluorescence (MPF) (Figure 4). In 2018, the team conducted intraoperative imaging of human breast tissue using its customized portable nonlinear optical imaging system to process the third harmonic (THG) to generate sEVs distribution images using its custom segmentation algorithm. They confirmed the higher density distribution of sEVs in highly graded tumor tissue and suggested the potential of sEVs as a label-free biomarker for cancer diagnosis and prognosis.119 Later, the team further investigated the label-free imaging of sEVs, which was accomplished by using multiphoton for direct visualization, characterization, and kinetic analysis of sEVs of breast cancer cells in different states based on their enriched NAD(P)H.120 Since MPF can only image auto-fluorescent material, but most of the chemical components in sEVs are non-fluorescent, imaging with MPF may suffer from data loss. Thus, the team introduced coherent anti-Stokes Raman scattering (CARS) to provide a more detailed chemical composition analysis. The team performed MPF and CARS imaging of rat mammary glands and performed K-means clustering analysis of CARS under the visualization and guidance of MPF to obtain more chemical information to classify the seen sEVs. They screened a group of sEVs suspected to be associated with cancer, which was validated in tissue images of human mammary tumors.121 The team also introduced fluorescence lifetime imaging microscopy (FLIM) for imaging sEVs, as FLIM can analyze the NAD(P)H status within sEVs and differentiate NADPH from NADP. It validated this experimentally and found that FLIM has the potential for imaging and characterizing sEVs to assess sEVs heterogeneity and to reflect cellular metabolic status by analyzing sEVs.122
 |
Figure 4 (A) Determination of phase of tumor cell invasion around desmoplasia by EVs distribution. (B) Intraoperative label-free multimodal imaging system. (A) and (B) Reprinted from Sun Y, You S, Tu H, et al. Intraoperative visualization of the tumor microenvironment and quantification of extracellular vesicles by label-free nonlinear imaging. Sci Adv. 2018. Creative Commons.119 (C)In vivo visualization of EVs from cancer tissue in rat mammary tumors by label-free multiphoton microscopy. Reprinted from You S, Barkalifa R, Chaney EJ, et al. Label-free visualization and characterization of extracellular vesicles in breast cancer. Proc Natl Acad Sci U S A. 2019;116(48):24012–24018. Creative Commons.120 (D) EVs cluster distribution in rat mammary tissues by K-means clustering of CARS spectra. Reprinted with permission from © The Optical Society. Sun Y, Chen EW, Thomas J, Liu Y, Tu H, Boppart SA. K-means clustering of coherent Raman spectra from extracellular vesicles visualized by label-free multiphoton imaging. Opt Lett. 2020;45(13):3613–3616.121 |
Summary and Outlook
All known methods of sEVs labeling have different advantages and disadvantages, the scope of application and limitations due to their different markers. Fluorescent dye labeling, as the most classical sEVs labeling technique, has the advantages of simple operation and low cost. However, based on the limitations of the current sEVs separation technology, it cannot effectively separate sEVs and small particles of lipids and proteins, while some lipophilic dyes will self-aggregate, which leads to unavoidable non-specific signals. At the same time, the influence of larger dye molecules on sEVs function, the instability of dye molecules, background fluorescence, and other drawbacks leave much room for optimization. Fluorescent dyes based on covalent binding for staining have increased specificity compared to common fluorescent dyes due to the relatively specific covalent binding reaction. Current click chemistry-based covalent binding labeling technology has dramatically improved the specificity and flexibility of labeling. Non-covalent conjugated labeling based on immunolabeling is another way to improve the flexibility and sensitivity of labeling, and the technical requirements and experimental costs are relatively low. However, the large number of antibodies and reporter molecules bound to the surface of sEVs will inevitably affect the physical properties and physiological functions of sEVs. Therefore, non-covalent binding labeling techniques based on smaller molecules such as aptamers, aptamer ligands, anchoring peptides, and electrostatic interactions have also been developed and applied. However, the use of these techniques is still at an initial stage. Genetic engineering-based fluorescent protein and luciferase labeling technology are more efficient, specific, flexible, and safe. However, it is relatively difficult to construct cell lines that express the correct stable and efficient fusion proteins.
For simple sEVs labeling and tracking experiments, traditional fluorescent dyes may be the most cost-effective and convenient choice. However, it should be noted that the use of PKH dyes for sEVs labeling should be avoided for its adverse performance, and the R18 dyes for sEVs labeling could be used to observe sEVs membrane fusion. If experimental cells are highly sensitive to dye toxicity or high imaging quality is required, attention can be directed towards newly developed novel dyes, some of which are commercially available. Immunolabeling techniques based on antigen–antibody reactions are another widely used fluorescence labeling method. They offer higher specificity compared to dye-based labeling methods, but staining efficiency is relatively lower. These techniques can be used for further subtyping and identification of sEVs based on choosing different antibody targets. However, immunolabeling should be avoided in experiments focusing on sEVs functionality due to its potential impact. If fluorescently labeled sEVs are required for long-term and large-scale experiments, biotechnological approaches may be a better choice. The characteristics and application of these fluorescent labeling techniques are summarized in Table 1.
 |
Table 1 Fluorescent Labeling Techniques of sEVs Imaging |
Labeling techniques based on radioisotope, nanoparticle, and chemical developers are more suitable for studying sEVs biodistribution. Their high sensitivity and resolution make them more suitable for deep tissue and in vivo tracer imaging. Thus, they have more greater clinical research value. However, the high research threshold of these labeling techniques, such as the use of isotopes, synthesis of nanoparticles, complex experimental operations, and the use of infrared imagers, photoacoustic imagers, MRI, and CT (even SPECT and PETCT), make it difficult to popularize these labeling techniques in some small and medium-sized laboratories. If qualifications and equipment allow, radioisotope labeling can achieve excellent imaging results and eliminate complex material synthesis steps. However, it requires the selection of suitable labeling media. For experiments involving sEVs-targeted therapy, AU nanoparticles with PTT effects and SPIONs, with excellent drug loading capabilities are excellent choices. Since different labeled imaging techniques have their own advantages and disadvantages, polymorphic imaging based on synthetic nanoparticles is a major research direction at present, and comprehensive analysis of information on the advantages of various imaging techniques can more effectively explore factual information on the processes of sEVs formation, release, circulation, uptake, and delivery of contents. All the aforementioned exogenous labeling methods have varying degrees of impact on sEVs. Therefore, label-free imaging techniques that have zero impact on sEVs hold great research prospects. These techniques, which require no labeling procedures, are extremely convenient and efficient. They enable in-situ detection of sEVs, leading to unparalleled advancements in clinical diagnostics. However, current research as a field is in its infancy, and the widespread adoption and dissemination of these technologies pose significant challenges. The characteristics and application of these non-fluorescent labeling techniques are summarized in Table 2.
 |
Table 2 Non-Fluorescent Labeling Techniques of sEVs Imaging |
With the advancement of sEVs research, the stability of labeling techniques and their impact on the biological function of sEVs have gained increasing attention. The stability of sEVs labeling is crucial for maintaining their traceability and detectability. Key factors include selecting stable labeling agents, appropriate labeling methods, and optimization of labeling conditions. For fluorescence labeling, fluorescence quenching is a significant factor to consider. Therefore, choosing dyes with high photostability can enhance the stability of labeling. Additionally, direct conjugation of fluorescent dyes can lead to dye transfer, which affects signal interpretation and weakens the fluorescence signal. Thus, using covalent or non-covalent labeling methods can improve labeling stability. Radioisotopes with poor biocompatibility require suitable chelators for selective labeling of the sEVs membrane or its contents, because direct labeling with radioisotopes has low efficiency and poor stability. Metal nanoparticles have improved their biocompatibility through materials engineering approaches. They are usually internalized into sEVs through donor cell uptake for labeling purposes. Regarding the impact on exosome function, membrane labeling is a simple and efficient method. However, the membrane plays a crucial role in the physiological function of sEVs and excessive labeling modifications on the membrane, including using large dye molecules, hierarchical connections of antigen–antibody complexes, or chelation of particles on the membrane surface, can significantly affect their function. Therefore, surface labeling techniques based on smaller molecules and intraluminal labeling techniques may be preferable for functional studies. It is recommended to perform functional assessments after sEVs labeling to evaluate the potential impact of labeling on their functionality. This can be done through nanoparticle tracking analysis, cellular uptake experiments, protein analysis, or functional assays to assess the uptake capacity, membrane fusion ability, and biological activity of labeled sEVs.
As a hot research topic in recent years, sEVs research is undoubtedly promising, and labeling and tracer imaging techniques provide the necessary technical support for sEVs-related studies in vivo and in vitro. However, various labeling techniques have their own advantages and disadvantages, and the appropriate labeling strategy should be chosen according to the experimental purpose. The development of marker imaging technology also advances our understanding of sEVs and makes them ultimately useful for clinical purposes.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [32070182]; Key Project of Jiangsu Commission of Health [ZD2021049] and High-level Talent Program at Affiliated Kunshan Hospital of Jiangsu University [gccrc2022002].
Disclosure
The authors report no potential conflicts of interest in this work.
References
1. Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750.
2. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. doi:10.1146/annurev-biochem-013118-111902
3. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659.
4. van Niel G, Carter DRF, Clayton A, Lambert DW, Raposo G, Vader P. Challenges and directions in studying cell-cell communication by extracellular vesicles. Nat Rev Mol Cell Biol. 2022;23(5):369–382. doi:10.1038/s41580-022-00460-3
5. Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8(7):727.
6. Yu S, Zhao Z, Xu X, Li M, Li P. Characterization of three different types of extracellular vesicles and their impact on bacterial growth. Food Chem. 2019;272:372–378. doi:10.1016/j.foodchem.2018.08.059
7. Murdica V, Giacomini E, Makieva S, et al. In vitro cultured human endometrial cells release extracellular vesicles that can be uptaken by spermatozoa. Sci Rep. 2020;10(1):8856. doi:10.1038/s41598-020-65517-9
8. Cook RL, Householder KT, Chung EP, Prakapenka AV, DiPerna DM, Sirianni RW. A critical evaluation of drug delivery from ligand modified nanoparticles: confounding small molecule distribution and efficacy in the central nervous system. J Control Release. 2015;220(Pt A):89–97. doi:10.1016/j.jconrel.2015.10.013
9. Santelices J, Ou M, Hui WW, Maegawa GHB, Edelmann MJ. Fluorescent Labeling of Small Extracellular Vesicles (EVs) Isolated from Conditioned Media. Bio-Protocol. 2022;12(12):76.
10. Wen S, Dooner M, Papa E, et al. Biodistribution of mesenchymal stem cell-derived extracellular vesicles in a radiation injury bone marrow murine model. Int J Mol Sci. 2019;20(21). doi:10.3390/ijms20215468
11. Wolf T, Baier SR, Zempleni J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma Caco-2 cells and rat small intestinal IEC-6 Cells. J Nutr. 2015;145(10):2201–2206. doi:10.3945/jn.115.218586
12. HP K, inventor; Viable cell labelling. US patent US4783401(A). patent application US19860925192. 1986.
13. Reclusa P, Verstraelen P, Taverna S, et al. Improving extracellular vesicles visualization: from static to motion. Sci Rep. 2020;10(1):6494. doi:10.1038/s41598-020-62920-0
14. Li Q, Li B, Li Q, et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018;9(9):854. doi:10.1038/s41419-018-0928-8
15. Pollalis D, Kim D, Nair GKG, Kang C, Nanda AV, Lee SY. Intraocular RGD-Engineered Exosomes and Active Targeting of Choroidal Neovascularization (CNV). Cells. 2022;11(16):2573.
16. Dehghani M, Gulvin SM, Flax J, Gaborski TR. Systematic Evaluation of PKH Labelling on Extracellular Vesicle Size by Nanoparticle Tracking Analysis. Sci Rep. 2020;10(1):9533. doi:10.1038/s41598-020-66434-7
17. Melling GE, Conlon R, Pantazi P, et al. Confocal microscopy analysis reveals that only a small proportion of extracellular vesicles are successfully labelled with commonly utilised staining methods. Sci Rep. 2022;12(1):262. doi:10.1038/s41598-021-04225-4
18. Shimomura T, Seino R, Umezaki K, et al. New lipophilic fluorescent dyes for labeling extracellular vesicles: characterization and monitoring of cellular uptake. Bioconjug Chem. 2021;32(4):680–684. doi:10.1021/acs.bioconjchem.1c00068
19. El-Andaloussi S, Lee Y, Lakhal-Littleton S, et al. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc. 2012;7(12):2112–2126. doi:10.1038/nprot.2012.131
20. Collot M, Ashokkumar P, Anton H, et al. MemBright: a family of fluorescent membrane probes for advanced cellular imaging and neuroscience. Cell Chem Biol. 2019;26(4):600–614 e7. doi:10.1016/j.chembiol.2019.01.009
21. Hyenne V, Ghoroghi S, Collot M, et al. Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. Dev Cell. 2019;48(4):554–572 e7. doi:10.1016/j.devcel.2019.01.014
22. Boyer MJ, Kimura Y, Akiyama T, et al. Endothelial cell-derived extracellular vesicles alter vascular smooth muscle cell phenotype through high-mobility group box proteins. J Extracell Vesicles. 2020;9(1):1781427. doi:10.1080/20013078.2020.1781427
23. Verderio C, Muzio L, Turola E, et al. Myeloid microvesicles are a marker and therapeutic target for neuroinflammation. Ann Neurol. 2012;72(4):610–624. doi:10.1002/ana.23627
24. Hu S, Morrin H, Wynne C, Meaney S. 3-Hexanoyl-7-nitrobenz-2-oxa-1,3-diazol-4-yl-cholesterol (3-NBD-cholesterol) is a versatile cholesterol tracer. Steroids. 2021;171:108840. doi:10.1016/j.steroids.2021.108840
25. Sharon N. Lectins: carbohydrate-specific reagents and biological recognition molecules. J Biol Chem. 2007;282(5):2753–2764. doi:10.1074/jbc.X600004200
26. Lennon KM, Wakefield DL, Maddox AL, et al. Single molecule characterization of individual extracellular vesicles from pancreatic cancer. J Extracell Vesicles. 2019;8(1):1685634. doi:10.1080/20013078.2019.1685634
27. Tian T, Zhu Y-L, Hu F-H, Wang -Y-Y, Huang N-P, Xiao Z-D. Dynamics of exosome internalization and trafficking. J Cell Physiol. 2013;228(7):1487–1495. doi:10.1002/jcp.24304
28. Gupta D, Liang X, Pavlova S, et al. Quantification of extracellular vesicles and using sensitive bioluminescence imaging. J Extracell Vesicles. 2020;9(1):1800222. doi:10.1080/20013078.2020.1800222
29. Coelho C, Vij R, Smith DQ, Brady NR, Hamacher-Brady A, Casadevall A. Study of Microbial Extracellular Vesicles:Separation by Density Gradients, Protection Assays and Labelling for Live Tracking. Bio Protoc. 2020;10(2):e3502. doi:10.21769/BioProtoc.3502
30. Puzar Dominkus P, Stenovec M, Sitar S, et al. PKH26 labeling of extracellular vesicles: characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim Biophys Acta Biomembr. 2018;1860(6):1350–1361. doi:10.1016/j.bbamem.2018.03.013
31. Rautaniemi K, Zini J, Lofman E, et al. Addressing challenges in the removal of unbound dye from passively labelled extracellular vesicles. Nanoscale Adv. 2021;4(1):226–240. doi:10.1039/d1na00755f
32. Takov K, Yellon DM, Davidson SM. Confounding factors in vesicle uptake studies using fluorescent lipophilic membrane dyes. J Extracell Vesicles. 2017;6(1):1388731. doi:10.1080/20013078.2017.1388731
33. Verweij FJ, Balaj L, Boulanger CM, et al. The power of imaging to understand extracellular vesicle biology in vivo. Nat Methods. 2021;18(9):1013–1026. doi:10.1038/s41592-021-01206-3
34. Dehghani M, Gaborski TR. Fluorescent labeling of extracellular vesicles. Methods Enzymol. 2020;645:15–42. doi:10.1016/bs.mie.2020.09.002
35. Radnaa E, Richardson LS, Sheller-Miller S, et al. Extracellular vesicle mediated feto-maternal HMGB1 signaling induces preterm birth. Lab Chip. 2021;21(10):1956–1973. doi:10.1039/d0lc01323d
36. Han J, Burgess K. Fluorescent indicators for intracellular pH. Chem Rev. 2010;110(5):2709–2728. doi:10.1021/cr900249z
37. Zhou X, Zhang J, Song Z, et al. ExoTracker: a low-pH-activatable fluorescent probe for labeling exosomes and monitoring endocytosis and trafficking. Chem Commun (Camb). 2020;56(94):14869–14872. doi:10.1039/d0cc06208a
38. Tian T, Wang Y, Wang H, Zhu Z, Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J Cell Biochem. 2010;111(2):488–496. doi:10.1002/jcb.22733
39. Di H, Zeng E, Zhang P, et al. General Approach to Engineering Extracellular Vesicles for Biomedical Analysis. Anal Chem. 2019;91(20):12752–12759. doi:10.1021/acs.analchem.9b02268
40. Roberts-Dalton HD, Cocks A, Falcon-Perez JM, et al. Fluorescence labelling of extracellular vesicles using a novel thiol-based strategy for quantitative analysis of cellular delivery and intracellular traffic. Nanoscale. 2017;9(36):13693–13706. doi:10.1039/c7nr04128d
41. Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev. 2010;39(4):1272–1279.
42. Xu L, Faruqu FN, Liam-Or R, et al. Design of experiment (DoE)-driven and uptake studies of exosomes for pancreatic cancer delivery enabled by copper-free click chemistry-based labelling. J Extracell Vesicles. 2020;9(1):1779458. doi:10.1080/20013078.2020.1779458
43. Wang M, Altinoglu S, Takeda YS, Xu Q. Integrating Protein Engineering and Bioorthogonal Click Conjugation for Extracellular Vesicle Modulation and Intracellular Delivery. PLoS One. 2015;10(11):e0141860. doi:10.1371/journal.pone.0141860
44. Zhu L, Xu Y, Wei X, et al. Coupling Aptamer-based Protein Tagging with Metabolic Glycan Labeling for In Situ Visualization and Biological Function Study of Exosomal Protein-Specific Glycosylation. Angew Chem Int Ed Engl. 2021;60(33):18111–18115. doi:10.1002/anie.202103696
45. Li N, Zhang W, Lin L, Shah SNA, Li Y, Lin JM. Nongenetically Encoded and Erasable Imaging Strategy for Receptor-Specific Glycans on Live Cells. Anal Chem. 2019;91(4):2600–2604. doi:10.1021/acs.analchem.8b05292
46. Chen C, Zong S, Wang Z, et al. Imaging and Intracellular Tracking of Cancer-Derived Exosomes Using Single-Molecule Localization-Based Super-Resolution Microscope. ACS Appl Mater Interfaces. 2016;8(39):25825–25833. doi:10.1021/acsami.6b09442
47. Mondal A, Ashiq KA, Phulpagar P, Singh DK, Shiras A. Effective Visualization and Easy Tracking of Extracellular Vesicles in Glioma Cells. Biol Proced Online. 2019;21:4. doi:10.1186/s12575-019-0092-2
48. Wan S, Zhang L, Wang S, et al. Molecular Recognition-Based DNA Nanoassemblies on the Surfaces of Nanosized Exosomes. J Am Chem Soc. 2017;139(15):5289–5292. doi:10.1021/jacs.7b00319
49. Gao X, Ran N, Dong X, et al. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci Transl Med. 2018;10:444.
50. Nimjee SM, White RR, Becker RC, Sullenger BA. Aptamers as Therapeutics. Annu Rev Pharmacol Toxicol. 2017;57:61–79. doi:10.1146/annurev-pharmtox-010716-104558
51. Huang M, Yang J, Wang T, et al. Homogeneous, Low-volume, Efficient, and Sensitive Quantitation of Circulating Exosomal PD-L1 for Cancer Diagnosis and Immunotherapy Response Prediction. Angewandte Chemie. 2020;59(12):4800–4805. doi:10.1002/anie.201916039
52. El Andaloussi S, Mäger I, Breakefield XO, Wood MJA. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. doi:10.1038/nrd3978
53. Akishiba M, Takeuchi T, Kawaguchi Y, et al. Cytosolic antibody delivery by lipid-sensitive endosomolytic peptide. Nat Chem. 2017;9(8):751–761. doi:10.1038/nchem.2779
54. Armstrong JPK, Holme MN, Stevens MM. Re-Engineering Extracellular Vesicles as Smart Nanoscale Therapeutics. ACS Nano. 2017;11(1):69–83. doi:10.1021/acsnano.6b07607
55. Cao H, Cheng Y, Gao H, et al. Tracking of Mesenchymal Stem Cell-Derived Extracellular Vesicles Improving Mitochondrial Function in Renal Ischemia-Reperfusion Injury. ACS Nano. 2020;14(4):4014–4026. doi:10.1021/acsnano.9b08207
56. Cao H, Yue Z, Gao H, et al. In Vivo Real-Time Imaging of Extracellular Vesicles in Liver Regeneration via Aggregation-Induced Emission Luminogens. ACS Nano. 2019;13(3):3522–3533. doi:10.1021/acsnano.8b09776
57. Meyer C, Losacco J, Stickney Z, Li L, Marriott G, Lu B. Pseudotyping exosomes for enhanced protein delivery in mammalian cells. Int J Nanomedicine. 2017;12:3153–3170. doi:10.2147/IJN.S133430
58. Men Y, Yelick J, Jin S, et al. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat Commun. 2019;10(1):4136. doi:10.1038/s41467-019-11534-w
59. Li TM, Lan WJ, Huang C, Zhang C, Liu XM. Establishment and identification of the near-infrared fluorescence labeled exosomes in breast cancer cell lines. Yi Chuan. 2016;38(5):427–435. doi:10.16288/j.yczz.16-017
60. Wachalska M, Rychlowski M, Grabowska K, et al. Palmitoylated mNeonGreen Protein as a Tool for Visualization and Uptake Studies of Extracellular Vesicles. Membranes. 2020;10(12). doi:10.3390/membranes10120373
61. Lai CP, Kim EY, Badr CE, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun. 2015;6:7029. doi:10.1038/ncomms8029
62. Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47(6):1118–1127. doi:10.1194/jlr.R600007-JLR200
63. Zhu L, Kalimuthu S, Gangadaran P, et al. Exosomes Derived From Natural Killer Cells Exert Therapeutic Effect in Melanoma. Theranostics. 2017;7(10):2732–2745. doi:10.7150/thno.18752
64. Takahashi Y, Nishikawa M, Takakura Y. In Vivo Tracking of Extracellular Vesicles in Mice Using Fusion Protein Comprising Lactadherin and Gaussia Luciferase. Methods Mol Biol. 2017;1660:245–254. doi:10.1007/978-1-4939-7253-1_20
65. Gangadaran P, Li XJ, Kalimuthu SK, et al. New Optical Imaging Reporter-labeled Anaplastic Thyroid Cancer-Derived Extracellular Vesicles as a Platform for In Vivo Tumor Targeting in a Mouse Model. Sci Rep. 2018;8(1):13509. doi:10.1038/s41598-018-31998-y
66. Luo W, Dai Y, Chen Z, Yue X, Andrade-Powell KC, Chang J. Spatial and temporal tracking of cardiac exosomes in mouse using a nano-luciferase-CD63 fusion protein. Commun Biol. 2020;3(1):114. doi:10.1038/s42003-020-0830-7
67. Huang A, Liu Y, Qi X, et al. Intravenously transplanted mesenchymal stromal cells: a new endocrine reservoir for cardioprotection. Stem Cell Res Ther. 2022;13(1):253. doi:10.1186/s13287-022-02922-z
68. Leng L, Wang Y, He N, et al. Molecular imaging for assessment of mesenchymal stem cells mediated breast cancer therapy. Biomaterials. 2014;35(19):5162–5170. doi:10.1016/j.biomaterials.2014.03.014
69. Corso G, Heusermann W, Trojer D, et al. Systematic characterization of extracellular vesicle sorting domains and quantification at the single molecule - single vesicle level by fluorescence correlation spectroscopy and single particle imaging. J Extracell Vesicles. 2019;8(1):1663043. doi:10.1080/20013078.2019.1663043
70. Rashid MH, Borin TF, Ara R, et al. Differential in vivo biodistribution of (131)I-labeled exosomes from diverse cellular origins and its implication for theranostic application. Nanomedicine. 2019;21:102072. doi:10.1016/j.nano.2019.102072
71. Morishita M, Takahashi Y, Nishikawa M, et al. Quantitative analysis of tissue distribution of the B16BL6-derived exosomes using a streptavidin-lactadherin fusion protein and iodine-125-labeled biotin derivative after intravenous injection in mice. J Pharm Sci. 2015;104(2):705–713. doi:10.1002/jps.24251
72. Royo F, Cossio U, Ruiz de Angulo A, Llop J, Falcon-Perez JM. Modification of the glycosylation of extracellular vesicles alters their biodistribution in mice. Nanoscale. 2019;11(4):1531–1537. doi:10.1039/c8nr03900c
73. Faruqu FN, Wang JT, Xu L, et al. Membrane Radiolabelling of Exosomes for Comparative Biodistribution Analysis in Immunocompetent and Immunodeficient Mice - A Novel and Universal Approach. Theranostics. 2019;9(6):1666–1682. doi:10.7150/thno.27891
74. Gonzalez MI, Martin-Duque P, Desco M, Salinas B. Radioactive Labeling of Milk-Derived Exosomes with (99m)Tc and In Vivo Tracking by SPECT Imaging. Nanomaterials. 2020;10(6). doi:10.3390/nano10061062
75. Hwang DW, Choi H, Jang SC, et al. Noninvasive imaging of radiolabeled exosome-mimetic nanovesicle using (99m)Tc-HMPAO. Sci Rep. 2015;5:15636. doi:10.1038/srep15636
76. Varga Z, Gyurkó I, Pálóczi K, et al. Radiolabeling of Extracellular Vesicles with (99m)Tc for Quantitative In Vivo Imaging Studies. Cancer Biother Radiopharm. 2016;31(5):168–173. doi:10.1089/cbr.2016.2009
77. Molavipordanjani S, Khodashenas S, Abedi SM, Moghadam MF, Mardanshahi A, Hosseinimehr SJ. (99m)Tc-radiolabeled HER2 targeted exosome for tumor imaging. Eur J Pharm Sci. 2020;148:105312. doi:10.1016/j.ejps.2020.105312
78. Choi H, Kim MY, Kim DH, et al. Quantitative Biodistribution and Pharmacokinetics Study of GMP-Grade Exosomes Labeled with (89)Zr Radioisotope in Mice and Rats. Pharmaceutics. 2022;14(6). doi:10.3390/pharmaceutics14061118
79. Khan AA, Man F, Faruqu FN, et al. PET Imaging of Small Extracellular Vesicles via [(89)Zr]Zr(oxinate)4 Direct Radiolabeling. Bioconjug Chem. 2022;33(3):473–485. doi:10.1021/acs.bioconjchem.1c00597
80. Jung KO, Kim Y-H, Chung S-J, et al. Identification of Lymphatic and Hematogenous Routes of Rapidly Labeled Radioactive and Fluorescent Exosomes through Highly Sensitive Multimodal Imaging. Int J Mol Sci. 2020;21(21):7850.
81. Warashina S, Zouda M, Mohri K, et al. 64Cu-labeling of small extracellular vesicle surfaces via a cross-bridged macrocyclic chelator for pharmacokinetic study by positron emission tomography imaging. Int J Pharm. 2022;624:121968. doi:10.1016/j.ijpharm.2022.121968
82. Horgan CC, Nagelkerke A, Whittaker TE, et al. Molecular imaging of extracellular vesicles in vitro via Raman metabolic labelling. J Mater Chem B. 2020;8(20):4447–4459. doi:10.1039/d0tb00620c
83. Betzer O, Perets N, Angel A, et al. In Vivo Neuroimaging of Exosomes Using Gold Nanoparticles. ACS Nano. 2017;11(11):10883–10893. doi:10.1021/acsnano.7b04495
84. Perets N, Betzer O, Shapira R, et al. Golden Exosomes Selectively Target Brain Pathologies in Neurodegenerative and Neurodevelopmental Disorders. Nano Lett. 2019;19(6):3422–3431. doi:10.1021/acs.nanolett.8b04148
85. Guo S, Perets N, Betzer O, et al. Intranasal Delivery of Mesenchymal Stem Cell Derived Exosomes Loaded with Phosphatase and Tensin Homolog siRNA Repairs Complete Spinal Cord Injury. ACS Nano. 2019;13(9):10015–10028. doi:10.1021/acsnano.9b01892
86. Lara P, Palma-Florez S, Salas-Huenuleo E, et al. Gold nanoparticle based double-labeling of melanoma extracellular vesicles to determine the specificity of uptake by cells and preferential accumulation in small metastatic lung tumors. J Nanobiotechnology. 2020;18(1):20. doi:10.1186/s12951-020-0573-0
87. Reguera J, Jiménez de Aberasturi D, Henriksen-Lacey M, et al. Janus plasmonic-magnetic gold-iron oxide nanoparticles as contrast agents for multimodal imaging. Nanoscale. 2017;9(27):9467–9480. doi:10.1039/c7nr01406f
88. Zhu D, Lyu M, Huang Q, et al. Stellate Plasmonic Exosomes for Penetrative Targeting Tumor NIR-II Thermo-Radiotherapy. ACS Appl Mater Interfaces. 2020;12(33):36928–36937. doi:10.1021/acsami.0c09969
89. Laurent S, Saei AA, Behzadi S, Panahifar A, Mahmoudi M. Superparamagnetic iron oxide nanoparticles for delivery of therapeutic agents: opportunities and challenges. Expert Opin Drug Deliv. 2014;11(9):1449–1470. doi:10.1517/17425247.2014.924501
90. Santhosh PB, Ulrih NP. Multifunctional superparamagnetic iron oxide nanoparticles: promising tools in cancer theranostics. Cancer Lett. 2013;336(1):8.
91. Zhuo Z, Wang J, Luo Y, et al. Targeted extracellular vesicle delivery systems employing superparamagnetic iron oxide nanoparticles. Acta Biomater. 2021;134:13–31. doi:10.1016/j.actbio.2021.07.027
92. Busato A, Bonafede R, Bontempi P, et al. Labeling and magnetic resonance imaging of exosomes isolated from adipose stem cells. Curr Protoc Cell Biol. 2017:75. doi:10.1002/cpcb.23
93. Busato A, Bonafede R, Bontempi P, et al. Magnetic resonance imaging of ultrasmall superparamagnetic iron oxide-labeled exosomes from stem cells: a new method to obtain labeled exosomes. Int J Nanomedicine. 2016;11:2481–2490. doi:10.2147/IJN.S104152
94. Jia G, Han Y, An Y, et al. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials. 2018;178:302–316. doi:10.1016/j.biomaterials.2018.06.029
95. Zhang F. Experimental Study on Construction of Nerve-Targeted MR Probe RVG-Exo-SPION Based on Exosomes. Master. Nanjing Medical University; 2020.
96. Pradhan P, Giri J, Banerjee R, Bellare J, Bahadur D. Preparation and characterization of manganese ferrite-based magnetic liposomes for hyperthermia treatment of cancer. J Magn Magn Mater. 2007;311(1):208–215. doi:10.1016/j.jmmm.2006.10.1179
97. Zhao M, Zhuang H, Zhang H, et al. A LRET Nanoplatform Consisting of Lanthanide and Amorphous Manganese Oxide for NIR-II Luminescence Lifetime Imaging of Tumor Redox Status. Angewandte Chemie. 2022:e202209592. doi:10.1002/anie.202209592
98. Abello J, Nguyen TDT, Marasini R, Aryal S, Weiss ML. Biodistribution of gadolinium- and near infrared-labeled human umbilical cord mesenchymal stromal cell-derived exosomes in tumor bearing mice. Theranostics. 2019;9(8):2325–2345. doi:10.7150/thno.30030
99. Jarockyte G, Daugelaite E, Stasys M, et al. Accumulation and toxicity of superparamagnetic iron oxide nanoparticles in cells and experimental animals. Int J Mol Sci. 2016;17(8):67.
100. Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4(6):435–446.
101. Michalet X, Pinaud FF, Bentolila LA, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307(5709):538–544. doi:10.1126/science.1104274
102. Dobhal G, Ayupova D, Laufersky G, Ayed Z, Nann T, Goreham RV. Cadmium-free quantum dots as fluorescent labels for exosomes. Sensors. 2018;18:87.
103. Zong S, Zong J, Chen C, et al. Single molecule localization imaging of exosomes using blinking silicon quantum dots. Nanotechnology. 2018;29(6):065705. doi:10.1088/1361-6528/aaa375
104. Jiang X, Zong S, Chen C, Zhang Y, Wang Z, Cui Y. Gold-carbon dots for the intracellular imaging of cancer-derived exosomes. Nanotechnology. 2018;29(17):175701. doi:10.1088/1361-6528/aaaf14
105. Rodrigues M, Richards N, Ning B, Lyon CJ, Hu TY. Rapid lipid-based approach for normalization of quantum-dot-detected biomarker expression on extracellular vesicles in complex biological samples. Nano Lett. 2019;19(11):7623–7631. doi:10.1021/acs.nanolett.9b02232
106. Zhang M, Vojtech L, Ye Z, Hladik F, Nance E. Quantum Dot Labeling and Visualization of Extracellular Vesicles. ACS Appl Nano Mater. 2020;3(7):7211–7222. doi:10.1021/acsanm.0c01553
107. Zhao J-Y, Chen G, Y-P G, et al. Ultrasmall Magnetically Engineered Ag2Se Quantum Dots for Instant Efficient Labeling and Whole-Body High-Resolution Multimodal Real-Time Tracking of Cell-Derived Microvesicles. J Am Chem Soc. 2016;138(6):1893–1903. doi:10.1021/jacs.5b10340
108. Bose RJC, Uday Kumar S, Zeng Y, et al. Tumor Cell-Derived Extracellular Vesicle-Coated Nanocarriers: an Efficient Theranostic Platform for the Cancer-Specific Delivery of Anti-miR-21 and Imaging Agents. ACS Nano. 2018;12(11):10817–10832. doi:10.1021/acsnano.8b02587
109. Shaikh S, Rehman FU, Du T, et al. Real-time multimodal bioimaging of cancer cells and exosomes through biosynthesized iridium and iron nanoclusters. ACS Appl Mater Interfaces. 2018;10(31):26056–26063. doi:10.1021/acsami.8b08975
110. Cao Y, Wu T, Zhang K, et al. Engineered Exosome-Mediated Near-Infrared-II Region VC Quantum Dot Delivery for Nucleus-Target Low-Temperature Photothermal Therapy. ACS Nano. 2019;13(2):1499–1510. doi:10.1021/acsnano.8b07224
111. Tayyaba RFU, Shaikh S, et al. In situ self-assembled Ag-FeO nanoclusters in exosomes for cancer diagnosis. J Materials Chem B. 2020;8(14):2845–2855. doi:10.1039/c9tb02610j
112. Ding Y-F, Li S, Liang L, et al. Highly biocompatible chlorin e6-loaded chitosan nanoparticles for improved photodynamic cancer therapy. ACS Appl Mater Interfaces. 2018;10(12):9980–9987. doi:10.1021/acsami.8b01522
113. Jang Y, Kim H, Yoon S, et al. Exosome-based photoacoustic imaging guided photodynamic and immunotherapy for the treatment of pancreatic cancer. J Control Release. 2021;330:293–304. doi:10.1016/j.jconrel.2020.12.039
114. Gilad AA, Ziv K, McMahon MT, van Zijl PCM, Neeman M, Bulte JWM. MRI reporter genes. J Nucl Med. 2008;49(12):1905–1908. doi:10.2967/jnumed.108.053520
115. Liu T, Zhu Y, Zhao R, Wei X, Xin X. Visualization of exosomes from mesenchymal stem cells in vivo by magnetic resonance imaging. Magn Reson Imaging. 2020;68:75–82. doi:10.1016/j.mri.2020.02.001
116. Liu T, Li Z, Li X, et al. In vivo visualization of murine melanoma cells B16-derived exosomes through magnetic resonance imaging. Biochimica Et Biophysica Acta General Subjects. 2022;1866(2):130062. doi:10.1016/j.bbagen.2021.130062
117. Imanbekova M, Suarasan S, Lu Y, Jurchuk S, Wachsmann-Hogiu S. Recent advances in optical label-free characterization of extracellular vesicles. Nanophotonics. 2022;11(12):2827–2863. doi:10.1515/nanoph-2022-0057
118. Shin H, Seo D, Choi Y. Extracellular vesicle identification using label-free surface-enhanced raman spectroscopy: detection and signal analysis strategies. Molecules. 2020;25(21). doi:10.3390/molecules25215209
119. Sun Y, You S, Tu H, et al. Intraoperative visualization of the tumor microenvironment and quantification of extracellular vesicles by label-free nonlinear imaging. Sci Adv. 2018;4(12):eaau5603. doi:10.1126/sciadv.aau5603
120. You S, Barkalifa R, Chaney EJ, et al. Label-free visualization and characterization of extracellular vesicles in breast cancer. Proc Natl Acad Sci U S A. 2019;116(48):24012–24018. doi:10.1073/pnas.1909243116
121. Sun Y, Chen EW, Thomas J, Liu Y, Tu H, Boppart SA. K-means clustering of coherent Raman spectra from extracellular vesicles visualized by label-free multiphoton imaging. Opt Lett. 2020;45(13):3613–3616. doi:10.1364/OL.395838
122. Sorrells JE, Martin EM, Aksamitiene E, et al. Label-free characterization of single extracellular vesicles using two-photon fluorescence lifetime imaging microscopy of NAD(P)H. Sci Rep. 2021;11(1):3308. doi:10.1038/s41598-020-80813-0
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
