Back to Journals » Infection and Drug Resistance » Volume 15
A Rapid Carbapenemase Genes Detection Method with Xpert Carba-R from Positive Blood Cultures Compared with NG-Test Carba 5 and Sequencing
Authors Xu Y, Song W , Huang P, Mei Y, Zhang Y, Xu T
Received 2 October 2022
Accepted for publication 13 December 2022
Published 28 December 2022 Volume 2022:15 Pages 7719—7725
DOI https://doi.org/10.2147/IDR.S392035
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yuqiao Xu,1,2,* Weijuan Song,1,2,* Peijun Huang,1,2 Yaning Mei,1,2 Yan Zhang,1,2 Ting Xu1,2
1Department of Laboratory Medicine, Jiangsu Province Hospital and Nanjing Medical University First Affiliated Hospital, Nanjing, People’s Republic of China; 2Branch of National Clinical Research Center for Laboratory Medicine, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yan Zhang; Ting Xu, Department of Laboratory Medicine, Jiangsu Province Hospital, Guangzhou Street No. 300, Nanjing, 210029, People’s Republic of China, Tel +8625-6830-6287, Fax +8625- 8372-4440, Email [email protected]; [email protected]
Objective: The objective of the current study was to evaluate the performance of Xpert Carba-R for the direct detection and identification of carbapenemase genes from positive blood cultures.
Methods: Pathogens which extracted from positive blood cultures and identified using MALDI-TOF MS as Enterobacterales were included in this study. Xpert Carba-R was used for the rapid detection of carbapenemase genes from positive blood cultures. NG-Test CARBA 5 and polymerase-chain reaction (PCR) sequencing were used for the detection of carbapenemases and carbapenemase genes in positive blood culture isolates, respectively. Finally, antibiotic susceptibility tests were conducted using the VITEK-2 Compact system.
Results: A total of 133 positive blood cultures of Enterobacterales were collected and 27 of them were detected to carry carbapenemase genes using Xpert Carba-R. In comparison with PCR sequencing results, the sensitivity and specificity of Xpert Carba-R and NG-Test CARBA 5 were calculated as 100%. Additionally, Xpert Carba-R could significantly shorten the turnaround time by directly detecting positive blood cultures comparing with NG-Test CARBA 5. For 27 carbapenem-producing strains, the resistance rates of carbapenems and aztreonam were 96.3% and 92.6%, respectively. Strains carrying the blaKPC gene were all sensitive to ceftazidime–avibactam. All strains were sensitive to tigecycline and colistin.
Conclusion: Xpert Carba-R is suitable for the rapid detection of main carbapenemase genes from positive blood cultures with high sensitivity and specificity. In comparison with NG-Test CARBA 5 and PCR sequencing methods, the timely and convenient method can be a useful test to guide optimal therapy and infection control.
Keywords: carbapenemase genes, positive blood culture, rapid detection, Xpert Carba-R
Introduction
In recent years, the emergence and widespread diffusion of antibiotic resistance has been a major public health concern, which can result in longer hospital stays, higher health care costs and mortality. It has been estimated that infections due to multidrug resistant bacteria can cause at least 700,000 deaths per year worldwide, and the number is expected to rise to 10 million by 2050.1 Multidrug resistant gram-negative bacteria can cause a variety of infections, including bloodstream infections (BSI), pneumonia and urinary tract infections with significant implications on antibiotic consumption and patients outcome. Among these infections, BSI is associated with high mortality. Bacteria invade the blood circulation, multiply in the blood, release toxins and metabolites, which can lead to systemic multiple organ dysfunction syndrome and even death.2
Approximately 71.5% of multidrug-resistant BSI pathogens are carbapenem-resistant Enterobacterales (CRE).3 Mechanisms of carbapenem resistance are heterogeneous, including carbapenemases production, extended-spectrum β-lactamases and/or AmpC cephalosporinases combined with altered membrane permeability. Among these, production of carbapenemases is the main resistance mechanism,4 and the rapid dissemination of carbapenem-producing Enterobacterales (CPE) throughout the world is worrisome and threatens public health. A report suggested that patients with CPE had approximately 4 times the odds of dying within 14 days compared to patients with non-CPE.5 Early detection of CPE infection is imperative for patient treatment, infection control and epidemiological studies.6
Various assays have been commercialized to detect carbapenemase activity or carbapenemase genes, including PCR sequencing, carbapenem hydrolysis assays, and colorimetric-based assays. Most of these assays detect carbapenemase activity and genes in bacterial colonies, and few have been evaluated directly on biological samples. Recently, the Xpert Carba-R assay, a PCR-based test run on the GeneXpert platform, was designed for the rapid detection and differentiation of 5 carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM, blaOXA-48) directly from clinical specimens.7 In previous studies, the sensitivity and specificity of the Xpert Carba-R assay were 100% and 98%, respectively.8 Published studies have shown that the Xpert Carba-R assay can be used to detect carbapenemase genes from strains,9 sputum,10 rectal swabs11 and bronchoalveolar lavage.12 In Aurélie Cointe’s study, Xpert Carba-R was shown to be suitable for the rapid detection of carbapenemase genes on positive blood vials by inoculating isolates of several variants in blood culture bottles. Limited numbers of clinical samples were evaluated in the study and all tests were negative.13
NG-Test Carba 5 is another newly developed method for detecting five major carbapenemases (KPC, NDM, IMP, VIM and OXA-48).14 Monoclonal antibodies were used for detection in bacterial colonies.
The aim of this study was to evaluate the clinical usefulness of Xpert Carba-R for the rapid detection of carbapenemase genes in positive blood cultures. The performance of the Xpert Carba-R assay and NG-Test CARBA 5 was compared for the detection of five carbapenemase genes or carbapenemases in positive blood culture broth or isolates.
Materials and Methods
Specimen Preparation
Blood cultures were processed according to routine methods using BD BACTEC™ FX blood culture system (Becton-Dickinson, Sparks, MD, USA). Once blood culture bottles were positive, gram staining was used for microscopic examination. Patients were included only one time when they happened BSI at the first time. If Gram stain showed a single form of gram-negative bacilli, pathogens would be enriched from positive blood culture fluid and directly identified by MALDI-TOF MS (BioMérieux, France). The specific methods for isolation and identification for pathogens were referred to the “in-house” MALI-TOF MS protocol for direct identification of GN bacteria from positive blood cultures described in the literature of Menglan Zhou et al.15 Enterobacterales isolated and identified in positive blood cultures were considered “potential carbapenemase producers” and included in the study. For positive blood cultures, isolates were inoculated onto blood agar plates (Antobiology, China). Blood agar plates were incubated at 35 °C overnight. To evaluate the accuracy of pathogens extracted from positive blood culture bottles, identification was carried out after colonies were grown on blood agar plates.
Xpert Carba-R
As blood cultures were positive, the Xpert Carba-R assay (Cepheid, Sunnyvale, CA, USA) was used to detect the carbapenemase genes in a cartridge. In brief, 40μL aliquot was directly mixed with Sample Reagent Buffer and vortexed. Then, 1.7mL of this sample reagent was transferred to an Xpert cartridge which was detected on the GeneXpert platform. Klebsiella pneumoniae ATCC BAA-1705 (blaKPC positive strain) and Escherichia coli ATCC 25922 (carbapenem-susceptible strain) were used as positive and negative controls.
PCR Sequencing Analysis
DNA was extracted from purified bacterial isolates using a TIANamp Bacteria DNA Kit (Tiagen, China) and tested by PCR with primers specific to five carbapenemase genes (blaKPC, blaNDM, blaVIM, blaOXA-48, blaIMP). A volume of 12.5μL of PCR Master Mix (Vazyme, USA) was mixed with 2μL of forward and reverse primers in a 25μL reaction. Reactions were amplified on the Applied Biosystems 2720 Thermal Cycler (ThermoFisher Scientific, USA) using following the cycling conditions: an initial 94℃ 2 min hold, followed by 36 cycles at 94℃ for 30s, 60℃ for 40s and 72℃ for 1 min, followed 72℃ for 5 min. The appropriately sized PCR products were confirmed by DNA sequence analysis.16 A positive result means that at least one carbapenemase gene was detected in this specimen. The negative result means that there were no blaKPC, blaNDM, blaVIM, blaOXA-48 or blaIMP carbapenemase genes detected by the PCR-based DNA sequence analysis. K. pneumoniae ATCC 2146 (blaNDM positive) and K. pneumoniae ATCC BAA-1705 (blaKPC positive) were used as positive controls. E. coli ATCC 25922 was used as a negative control.
NG-Test CARBA 5
Bacterial isolates were tested using the NG-Test CRABA 5 kit (Fosun Diagnostics, China) simultaneously. The NG-Test CRABA 5 is a qualitative rapid lateral flow assay with mouse monoclonal antibodies against KPC, NDM, IMP, VIM and OXA-48, which are immobilized on nitrocellulose membrane test zones. The suspected colony was mixed with 150 μL extraction buffer, and then 100 μL of this mixture was dispensed into the cassette well and allowed to migrate toward the conjugate pad. The carbapenemase-antibody complexes migrated through the nitrocellulose membrane and were captured by corresponding anti-carbapenemase monoclonal antibodies immobilized on the membrane, resulting in a red line (or lines) on the test zone(s) and on the control zone.
Antibiotic Sensitivity Test
Antibiotic susceptibility tests were conducted using the VITEK-2 Compact system (BioMérieux, France) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI 2020, version M100-S30). Ceftazidime–avibactam (Oxoid, England) were tested using Kirby–Bauer method.
Data Analysis and Statistics Analysis
We calculated the sensitivity and specificity of the Xpert Carba-R and NG-Test CARBA 5 when compared with the PCR sequencing. Antibiotic sensitivity analyses were performed using Whonet (version 5.6).
Results
A total of 300 blood vials were collected in Jiangsu Province Hospital from May 2020 to June 2021 (Figure 1). Through rapid identification, 133 positive blood cultures of Enterobacterales were detected by Xpert Carba-R for carbapenemase genes and 27 CPE were obtained (Table 1). The most common pathogenic bacteria identified among those CPE was K. pneumoniae (92.6%), followed by E. coli (3.7%) and Citrobacter freundii (3.7%). Using the Xpert Carba-R, the blaKPC gene was identified in 24 strains of K. pneumoniae, and the blaNDM gene was identified in 2 strains of K. pneumoniae, 1 strain of E. coli and 1 strain of C. freundii. The results of PCR sequencing and NG-Test CARBA 5 are also displayed in Table 1. All results obtained by using the commercial kits were consistent with the PCR sequencing results, and the sensitivity and specificity of Xpert Carba-R and the NG-Test CARBA 5 were calculated as 100%.
 |
Table 1 Results of the Xpert Carba-R and the NG-Test Carba 5 |
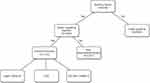 |
Figure 1 Detect flow of positive blood cultures. |
Table 2 shows the characteristics of Xpert Carba-R and NG-Test CARBA 5. The specimens of Xpert Carba-R were positive blood culture, so the setup time of Xpert Carba-R was only 3–5 min. But for NG-Test CARBA 5, the setup time was significantly increased to 16 h due to the second incubation required for the assay. As to the testing cost, the Xpert Carba-R was higher than the NG-Test CARBA 5.
 |
Table 2 Testing Parameters of the Xpert Carba-R and NG-Test CARBA 5 |
Results of antibiotic sensitivity tests are shown in Table 3. All 27 CPE isolates were resistant to cephalosporin, piperacillin–tazobactam and ampicillin–sulbactam. The resistance rates of carbapenem and aztreonam were 96.3% and 92.6%, respectively. Strains carrying blaKPC gene were sensitive to ceftazidime–avibactam. No isolates were resistant to tigecycline or colistin. Among the 27 CPE patients, 10 patients were treated with monotherapy including tigecycline, polymyxin B and carbapenems. The remaining 17 patients were treated with two or more antibiotics, and the main combination regimen was carbapenems combined with tigecycline or polymyxin B. When the patients were discharged from the hospital, 18 patients recovered, 9 patients did not recover and two patients were died in hospital.
 |
Table 3 Sensitivity and Resistance of 27 CPE |
Discussion
Since the early 2000s, CRE isolates have emerged worldwide and these isolates are mainly due to acquired carbapenemases.10 CRE infections have been a significant hospital threat and an emerging public-health problem. Reports from the China CRE Network showed that the overall CRE infection rates varied significantly by region, and the rate in Jiangsu was the highest.17 Approximately 32% of patients with BSI caused by CRE die within 14 days.5 Hence, early detection of patients carrying and/or CPE infection in positive blood cultures is essential.
Rapid methods, including Xpert Carba-R and Carba-NP18 have been proposed for the rapid detection of CPE from positive blood cultures. Carba-NP detect CPE basing on acid production during imipenem hydrolysis.19 Unfortunately, Carba-NP cannot detect the type of carbapenemase. It is capable detecting carbapenemase from Enterobacterale-positive blood cultures, but in cannot differentiate CPE expressing serine carbapenemases from those expressing metallo-β-lactamase.20 To date, two reports on rapid carbapenemase genes detection with Xpert Carba-R directly from positive blood cultures have been published in PubMed. Jaureguy et al reported good performance of this test on aerobic vials inoculated with isolates, but that did not contain blood.21 Aurélie Cointe et al focused on assessing the influence of the patient’s blood on the performance of the test, but only a few assays worked directly on positive blood cultures.13 In our study, the sensitivity and specificity of Xpert Carba-R were 100% compared with the PCR sequencing method. Of 133 positive blood cultures, 27 CPE were obtained. K. pneumoniae accounted for the majority of carbapenemase-producing isolates among Enterobacterales, which was the consistent with previous studies.17 Ninety-six percent of K. pneumoniae carried blaKPC gene, suggesting that blaKPC remains the main genotype of K. pneumoniae.
In our study, we compared two rapid tests for the five carbapenemase genes or carbapenemases, namely the Xpert Carba-R and NG-Test CARBA 5. Both of them showed excellent performance with the sensitivity and specificity of 100%. These two accurate assays can also shorten the testing time to within 2 h, but PCR sequencing needed 54–86 h. However, Xpert Carba-R could directly detect carbapenemase genes in positive vials, which could significantly reduce the setup time. No culture-based methods may reduce the time needed to get the antibiotic resistance data, leading to an earlier institution of effective antimicrobial treatment. In previous studies, neither assays could detect specific blaIMP subtypes,14 and no isolate producing IMP was detected in this study.
The results of antibiotic sensitivity tests showed that 27 CPE were all sensitive to tigecycline and colistin. Qureshi et al evaluated a cohort study on patients with CRE infection and the results showed that in patients who received combination therapy with carbapenem/tigecycline or carbapenem/colistin, 28-day survival was significantly higher than in those on monotherapy.22 Strains carrying blaKPC gene were sensitive to ceftazidime–avibactam. Ceftazidime–avibactam, a novel β-lactam combination, has been demonstrated to have a higher rate of clinical success than other antimicrobial regimens for BSI.23
This study has some limitations. First, our convenience sample size was relatively small due to our limited available resources for this study. However, this was the first attempt to study the performance of Xpert Carba-R in positive blood cultures from clinical samples. Another limitation may be due to the limited variety of carbapenemase genes. Due to local epidemiology, blaIMP, blaVIM and blaOXA-48 were not detected in our study.
Conclusion
Xpert Carba-R is suitable for the rapid detection of main carbapenemase genes from positive blood cultures with high sensitivity and specificity. In comparison with NG-Test CARBA 5 and PCR sequencing methods, the timely and convenient method can be a useful test to guide optimal therapy and infection control.
Ethics and Consent Statement
This work was approved by the Ethics Committee at Jiangsu Province Hospital and complied with the Declaration of Helsinki. Written informed consent and confidentiality agreements were obtained from the patient.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bassetti M, Vena A, Giacobbe DR, Castaldo N. Management of infections caused by multidrug-resistant gram-negative pathogens: recent advances and future directions. Arch Med Res. 2021;52(8):817–827. doi:10.1016/j.arcmed.2021.09.002
2. Zhang S, Yang Z, Sun L, et al. Clinical observation and prognostic analysis of patients with Klebsiella pneumoniae bloodstream infection. Front Cell Infect Microbiol. 2020;10:577244. doi:10.3389/fcimb.2020.577244
3. Li X, Ye H. Clinical and mortality risk factors in bloodstream infections with carbapenem-resistant Enterobacteriaceae. Can J Infect Dis Med Microbiol. 2017;2017:6212910. doi:10.1155/2017/6212910
4. Tilahun M, Kassa Y, Gedefie A, Ashagire M. Emerging carbapenem-resistant Enterobacteriaceae infection, its epidemiology and novel treatment options: a review. Infect Drug Resist. 2021;14:4363–4374. doi:10.2147/IDR.S337611
5. Tamma PD, Goodman KE, Harris AD, et al. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae Bacteremia. Clin Infect Dis. 2017;64(3):257–264. doi:10.1093/cid/ciw741
6. Humphries RM, McKinnell JA, Burnham C-AD. Continuing challenges for the clinical laboratory for detection of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2015;53(12):3712–3714. doi:10.1128/JCM.02668-15
7. Li HH, He ZJ, Xie LM, et al. Evaluation of Xpert Carba-R assay for the detection of carbapenemase genes in gram-negative bacteria. Biomed Res Int. 2021;2021:6614812. doi:10.1155/2021/6614812
8. Moubareck CA, Hammoudi HD, Sartawi M, Lawlor K, Sarkis DK, Alatoom A. Assessment of the performance of CHROMagar KPC and Xpert Carba-R assay for the detection of carbapenem-resistant bacteria in rectal swabs: first comparative study from Abu Dhabi, United Arab Emirates. J Glob Antimicrob Resist. 2020;20:147–152. doi:10.1016/j.jgar.2019.07.021
9. Zhou M, Kudinha T, Du B, et al. Active surveillance of Carbapenemase-Producing Organisms (CPO) Colonization With Xpert Carba-R assay plus positive patient isolation proves to be effective in CPO containment. Front Cell Infect Microbiol. 2019;9:162. doi:10.3389/fcimb.2019.00162
10. Cai Z, Tao J, Jia T, et al. Multicenter evaluation of the Xpert Carba-R assay for detection and identification of carbapenemase genes in sputum specimens. J Clin Microbiol. 2020;58(9). doi:10.1128/JCM.00644-20
11. Jin X, Zhang H, Wu S, et al. Multicenter evaluation of Xpert Carba-R assay for detection and identification of the carbapenemase genes in rectal swabs and clinical isolates. J Mol Diagn. 2021;23(1):111–119. doi:10.1016/j.jmoldx.2020.10.017
12. Vergara A, Moreno-Morales J, Roca I, et al. A comparative study between real-time PCR and loop-mediated isothermal amplification to detect carbapenemase and/or ESBL genes in Enterobacteriaceae directly from bronchoalveolar lavage fluid samples. J Antimicrob Chemother. 2020;75(6):1453–1457. doi:10.1093/jac/dkaa031
13. Cointe A, Walewski V, Hobson CA, et al. Rapid carbapenemase detection with Xpert Carba-R V2 directly on positive blood vials. Infect Drug Resist. 2019;12:3311–3316. doi:10.2147/IDR.S204436
14. Kanahashi T, Matsumura Y, Yamamoto M, Tanaka M, Nagao M. Comparison of the Xpert Carba-R and NG-Test CARBA5 for the detection of carbapenemases in an IMP-type carbapenemase endemic region in Japan. J Infect Chemother. 2021;27(3):503–506. doi:10.1016/j.jiac.2020.11.001
15. Zhou M, Yang Q, Kudinha T, et al. An improved in-house MALDI-TOF MS protocol for direct cost-effective identification of pathogens from blood cultures. Front Microbiol. 2017;8:1824. doi:10.3389/fmicb.2017.01824
16. Traczewski MM, Carretto E, Canton R, Moore NM, Richter SS. Multicenter evaluation of the Xpert Carba-R assay for detection of carbapenemase genes in gram-negative isolates. J Clin Microbiol. 2018;56(8). doi:10.1128/JCM.00272-18
17. Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE network. Antimicrob Agents Chemother. 2018;62(2). doi:10.1128/AAC.01882-17
18. Seco B, Campos JC, Da CRD, et al. Improved blood culture workflow for faster identification of KPC-producing Enterobacterales. Braz J Microbiol. 2019;50(1):127–132. doi:10.1007/s42770-018-0037-y
19. Fernandez J, Rodriguez-Lucas C, Fernandez-Suarez J, Vazquez F, Rodicio MR. Identification of Enterobacteriaceae and detection of carbapenemases from positive blood cultures by combination of MALDI-TOF MS and Carba NP performed after four hour subculture in Mueller Hinton. J Microbiol Methods. 2016;129:133–135. doi:10.1016/j.mimet.2016.08.014
20. Bianco G, Boattini M, Iannaccone M, Zanotto E, Cavallo R, Costa C. Direct ethylenediaminetetraacetic acid-modified beta-lactam inactivation method: an improved method to identify serine-carbapenemase-, Metallo-beta-Lactamase-, and extended-spectrum-beta-lactamase-producing enterobacterales directly from positive blood culture. Microb Drug Resist. 2021;27(6):740–746. doi:10.1089/mdr.2020.0343
21. Jaureguy F, Mansour H, Bigot J, et al. Use of the Xpert CarbaR assay for direct detection of carbapenemase genes from blood cultures and urine samples. J Hosp Infect. 2018;98(3):245–246. doi:10.1016/j.jhin.2017.09.026
22. Alhashem F, Tiren-Verbeet NL, Alp E, Doganay M. Treatment of sepsis: what is the antibiotic choice in bacteremia due to carbapenem resistant Enterobacteriaceae? World J Clin Cases. 2017;5(8):324–332. doi:10.12998/wjcc.v5.i8.324
23. Gaibani P, Re MC, Campoli C, Viale PL, Ambretti S. Bloodstream infection caused by KPC-producing Klebsiella pneumoniae resistant to ceftazidime/avibactam: epidemiology and genomic characterization. Clin Microbiol Infect. 2020;26(4):511–516. doi:10.1016/j.cmi.2019.11.011
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
