Back to Journals » Infection and Drug Resistance » Volume 14
A Rapid Antimicrobial Susceptibility Test for Klebsiella pneumoniae Using a Broth Micro-Dilution Combined with MALDI TOF MS
Authors Wang G , Song G , Xu Y
Received 4 February 2021
Accepted for publication 22 April 2021
Published 14 May 2021 Volume 2021:14 Pages 1823—1831
DOI https://doi.org/10.2147/IDR.S305280
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Gang Wang,1,* Guobin Song,1,2,* Yuanhong Xu1
1Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui Province, People’s Republic of China; 2Department of Clinical Laboratory, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yuanhong Xu
Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, 218 Jixi Road, Shushan District, Hefei, Anhui Province, 230022, People’s Republic of China
Email [email protected]
Background: Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a novel method that can be used to identify pathogens and has potential applications in the detection of drug-resistant bacteria.
Purpose: To evaluate the ability of a MALDI-TOF MS-based broth micro-dilution method in detecting the minimum inhibitory concentration (MIC) values of Klebsiella pneumoniae to ceftriaxone and imipenem.
Materials and Methods: Sixty strains of K. pneumoniae with different levels of resistance to carbapenems and cephalosporins were randomly collected. The 0.5 McFarland (Mc) concentration of the bacterial suspension was inoculated in cation-adjusted Mueller-Hinton broth (CAMHB) with a final cell turbidity of 5× 105 CFU/mL. The broth was incubated with serial concentrations of antibiotics. After centrifuging the bacterial suspensions, the lysed cells were analyzed by MALDI-TOF MS to identify the growth-promoting or inhibitory effects on K. pneumoniae. The molecular mechanisms of resistance were investigated by PCR and DNA sequencing analysis.
Results: The expression of known resistance genes (blaKPC, blaFOX, blaDHA, blaCTX-M and blaTEM) was detected in the 30 carbapenems-resistant strains. The agreement between the MIC values derived from the MALDI-TOF MS analysis and from the broth micro-dilution method was 61.7% for ceftriaxone and 71.7% for imipenem. According to the Clinical and Laboratory Standards Institute (CLSI) breakpoint of resistance to ceftriaxone and imipenem, the 60 isolates were accurately classified as resistant or susceptible isolates with 100% sensitivity and 100% specificity.
Conclusion: The transmission and infection of multidrug-resistant bacteria could be better managed and treated with the rapid identification of strains and antimicrobial susceptibility. A MALDI-TOF MS-based susceptibility test could be used to identify resistance of K. pneumoniae within a short time-frame. This approach could potentially be used as a supplementary antimicrobial susceptibility test that could be investigated on more bacterial species combined with different antibiotics.
Keywords: Klebsiella pneumoniae, matrix-assisted laser desorption ionization–time of flight mass spectrometry, carbapenemases, extended-spectrum β-lactamases, antimicrobial susceptibility test
Introduction
MALDI-TOF MS has become established as a robust analytical method that can be used to identify strains of bacteria and fungi, as it is accurate, inexpensive and has high throughput and a short turnaround time.1 For clinically important gram-negative bacteria, particularly intestinal microbes, such as E. coli, K. pneumoniae, Acinetobacter baumannii and P. aeruginosa, the direct cell smear method is commonly conducted to acquire mass spectra that are used for species identification with an accuracy of >98%. The formic acid extraction (on-plate) method is applied to lyse the gram-positive bacterial cell walls and yeast to increase the probability of accurate identification to >97%. However, the correct identification of filamentous fungi remains challenging even when using the ethanol-formic acid extraction method.2,3
Several studies have reported the direct identification of pathogens from blood and urine samples without prior culturing that could be used for rapid diagnosis.4–6 These innovations have resulted in a major decrease in the times required for species identification to several minutes compared to around one day using traditional biochemical identification. However, universal susceptibility testing to detect the impact of antibiotics on microorganisms still requires at least one day. The efficiency gap between microbial identification and drug susceptibility has driven the development of novel approaches for the rapid detection and identification of drug resistance in microorganisms. In this setting, MALDI-TOF MS has shown major potential in the detection of antimicrobial resistance and strain typing. MALDI-TOF MS approaches have been used to characterize patterns of antimicrobial resistance by measuring the molecular modifications of antibiotics due to the enzyme activities of pathogens.7,8 It has also been used to analyze the mass peak profiles of bacteria that correspond to the structures of enzymes and proteins related to drug resistance,9–11 and to measure the effects of antibiotics on the growth of microorganisms.12,13
The huge potential of MALDI-TOF MS in the rapid detection of drug resistance has been demonstrated by the enzyme-mediated antibiotics hydrolysis assays. The different genotypes of carbapenemase-producing K. pneumoniae from urine and bloodstream can be successfully discriminated within 3 h using the ertapenem and imipenem hydrolysis assays, and the results showed 100% agreement with genotypic characterization.14,15 Also, ESBLs-producing Enterobacteriaceae can be detected by observing the mass peaks shifts of ceftriaxone and cefotaxime.16 The successful establishment of mass spectra models correlating to specific mechanisms of resistance could expedite the direct detection of resistance when performing species identification. This could also include applications such as the direct differentiation of MRSA and MSSA, VIM and SPM-type β-lactamase-producing Pseudomonas aeruginosa.10,17
Several other studies have proposed a new method called the MALDI Biotyper-Antibiotic Susceptibility Test Rapid Assay (MBT-ASTRA) to differentiate antimicrobial-resistant and sensitive strains in a short time of 2–4 h.18–20 MBT-ASTRA experiments have used a relative growth (RG) algorithm to calculate the ratio of area under the curves (AUCs) for bacteria mass spectra when the test strains were incubated with and without antibiotics. The approach allowed the quantitative detection of bacteria growth under different concentrations of antibiotics using MALDI-TOF MS. Also, an important aspect of this method allows the resistance and susceptibility of different bacterial strains to be distinguished at applicable breakpoint concentrations of various antibiotics.
Direct-on-Target Micro-droplet Growth Assay (DOT-MGA) is another MALDI-TOF MS-based method that can be used to rapidly detect resistant and sensitive strains. This approach directly uses the MS plate to support the incubation of bacteria with and without antibiotics in micro volumes of liquid broth.21,22 The main processes involve the culture of bacterial isolates with and without antibiotics in a volume of 2–10 μL of broth for an appropriate incubation time and the mass spectra analysis was performed directly after removing the liquids. The identification score was applied to interpret the results as resistant or susceptible strains. However, this assay can only differentiate resistant and sensitive strains, but the exact MIC values cannot be determined. Also, a new mass spectrometry-based method has been proposed by combining MBT-ASTRA and DOT-MGA for the rapid determination of MIC values.23
Klebsiella pneumoniae is one of the most common opportunistic pathogens found in the clinic. As a consequence of the over-use of cephalosporins, carbapenems-resistant K. pneumoniae (CRKP) has emerged making the treatment of multidrug resistance very challenging. The prevalence of CRKP varies across different populations, regions and countries from 16% to nearly 40%.24,25 There is an urgent need to diagnose CRKP strains and prevent their transmission in hospitals at the early stage of infection. In this study, we developed a MALDI-TOF MS-based assay that was compared to the reference broth micro-dilution method. Based on the resistance to carbapenems and cephalosporins mediated by CRKP strains, we evaluated the ability of the new method for the rapid detection of drug resistance to ceftriaxone and imipenem in K. pneumoniae strains.
Materials and Methods
Characterization of the Bacterial Strains
Sixty non-replicated strains of K. pneumoniae were randomly collected from October 2018 to May 2019 at the First Affiliated Hospital of Anhui Medical University. The isolates were divided into two groups specifically: carbapenems-resistant and multidrug-sensitive isolates. The antimicrobial susceptibility test was performed using the disc diffusion method and the VITEK-2 Compact system (BioMérieux, France). The procedure for the tests and the interpretation of resistant (R), intermediate (I), and susceptible (S) isolates followed the Clinical Laboratory Standards Institute Criteria (CLSI). The genotype of drug resistance was screened by PCR for carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM, blaSME, blaGES), ESBL genes (blaCTX-M, blaTEM, blaSHV) and AmpC genes (blaFOX, blaDHA, blaMOX, blaCIT, blaACC, blaEBC).26 Then, DNA sequencing was conducted by Sangon (Shanghai, China). The PCR results were validated by nucleotide BLAST searches using GenBank.
Antimicrobial Susceptibility Test by Broth Micro-Dilution Method
The operational process using a broth micro-dilution method was performed according to CLSI guidelines. Briefly, 0.5 Mc suspension of the isolates was prepared and diluted 100-fold with cation-adjusted Mueller-Hinton broth (CAMHB). 50 μL of the bacterial suspension was seeded into a 96-well plate containing 50 μL of CAMHB with serial concentrations of antibiotics. The final inoculum of the bacteria was approximately 5×105 CFU/mL. The final concentrations of imipenem and ceftriaxone were 128 μg/mL, 64 μg/mL, 32 μg/mL, 16 μg/mL, 8 μg/mL, 4 μg/mL, 2 μg/mL, 1 μg/mL, 0.5 μg/mL, 0.25 μg/mL and 0.125 μg/mL. The suspension was cultured at 35°C for 18–20 h. Antimicrobial susceptibility testing was performed in triplicate and the median MIC values were calculated.
Antimicrobial Susceptibility Test by MALDI-TOF MS
Each strain was tested in the same way as the broth micro-dilution method, with a shorter incubation time of 4 h at 35°C. Then, all the suspensions were centrifuged at 4000 rpm for 10 minutes to separate the bacterial cells. The supernatant was discarded and 5 μL of 70% formic acid was added to lyse the isolates. 2 μL of the bacterial lysate was spotted onto the MS plate, overlaid with 1 μL of CHCA (α-cyano-4-hydroxycinnamic acid) and allowed to dry at room temperature. The spectra were acquired using the RUO mode of the MALDI-TOF MS instrument (BioMérieux, France). The spectra was analyzed using the SARAMIS Premium software. The quality control strain of Escherichia coli ATCC8739 was used for calibration of the Vitek-MS system. Each strain was accompanied by a growth control inoculating isolate without antibiotics and a sterile control without isolates and antibiotics. Each strain test was performed in triplicate. The results were considered reliable and valid when the MICs were identical in three parallel tests.
The Interpretation of MIC Values Derived from the Broth Micro-Dilution Method and MALDI-TOF MS
The MIC results of the broth micro-dilution method (BMD-MIC) were defined as the lowest concentration of antibiotics that resulted in the complete inhibition of visible bacterial growth. The MS spectra obtained with different concentrations of antimicrobial agents are correlated with the growth of the strain. Identification scores ≥90 indicated successful growth detection of the strain and showed that the strain was not inhibited at certain concentrations. The MIC values determined by MALDI-TOF MS (MS-MIC) were the lowest concentrations at which the identification score was <90 or had no identification equivalent to the visible inhibition of bacterial growth.
Statistical Analysis
The feasibility of the MALDI-TOF MS-based MIC test was evaluated by simultaneously performing a broth micro-dilution method. The broth micro-dilution test is a reference method used to determine the MIC. These data obtained with MALDI-TOF MS were compared to the MIC results of the standard method. The levels of agreement in the MIC values determined by the two methods were calculated.
Results
The 60 collected isolates were screened for their susceptibility to antimicrobial agents using a clinic workflow with a VITEK-2 Compact system and the disc diffusion method. The 30 carbapenems-resistant strains showed multidrug resistance to most antibiotics except for tigecycline, polymyxin, minocycline and trimesulf, to which the susceptibilities were of 100%, 100%, 83% and 67%, respectively. The 30 sensitive-isolates did not carry carbapenemase genes, ESBL genes or AmpC genes. Amongst all the targeted carbapenemase genes, only the KPC-carbapenemase was detected in 29 isolates. These bacterial strains also carried at least one type of ESBL genes or AmpC genes including blaCTX-M1, blaCTX-M9, blaTEM, blaFOX and blaDHA. (as shown in Figure 1) One carbapenems-resistant strain did not express any carbapenemase genes but simultaneously expressed blaCTX-M1, blaFOX and blaDHA.
The incubation conditions were investigated with a suspension of 5×105 CFU/mL bacterial cells after culturing at 35°C for 1, 2, 3 and 4 h, respectively. The suspensions without antibiotics were successfully identified by MALDI-TOF MS with a score >90 after 4 h incubation. (shown in Figure 2) The subsequent experiment was conducted with a 4 h incubation before separating the bacterial cells and the broth. The MIC of ceftriaxone and imipenem detected using the VITEK-2 Compact system were ≥64 μg/mL and ≥16 μg/mL, respectively, in the resistant strains. The MIC values of the susceptible isolates were ≤1 μg/mL.
 |
Figure 2 The mass spectra of K. pneumoniae acquired by MALDI-TOF MS. Notes: (A) Successful identification with a score ≥90; (B) Failed identification with a score <90. |
The MIC values of ceftriaxone determined by the broth micro-dilution method were >128 μg/mL in the 29 resistance strains. The MIC value derived from the MALDI-TOF MS was 128 μg/mL in 22 of the 29 resistant isolates. The other eight resistant isolates reached the same values between the BMD-MIC and MS-MIC. In the sensitive isolates, only one had a lower MS-MIC than BMD-MS.
The BMD-MIC values of imipenem were 128 μg/mL in 28 resistant K. pneumoniae strains and 64 μg/mL in the other two isolates. Compared to BMD-MIC, 9 strains had one-concentration-gradient lower MIC values of 64 μg/mL detected by MALDI-TOF MS. The MS-MIC values of imipenem decreased by two-concentration gradients in four resistant isolates, of which two had a MIC of 32 μg/mL and two had a value of 16 μg/mL. The MS-MIC values of the four carbapenems-sensitive isolates were lower than the determined BMD-MIC, two decreased from 1 to 0.5 μg/mL, and the other two decreased from 0.5 to 0.25 μg/mL (as shown in Tables 1 and 2).
 |
Table 1 The MIC of Ceftriaxone and Imipenem Determined by MALDI-TOF MS and Broth Micro-Dilution Method in 30 Carbapenems-Resistant Strains of K. pneumoniae |
 |
Table 2 The MIC of Ceftriaxone and Imipenem Determined by MALDI-TOF MS and Broth Micro-Dilution Method in 30 Susceptible Strains of K. pneumoniae |
The ceftriaxone- and imipenem-resistant strains had lower MIC values detected by MALDI-TOF MS compared to those derived from the broth micro-dilution method, which accounted for 73.33% (22/30) and 43.33% (13/30) of the discrepancies, respectively. The susceptible strains showed a high concordance between the BMD-MIC and MS-MIC values that reached 96.67% (29/30) and 86.67% (26/30) for ceftriaxone and imipenem, respectively. The agreement between the MIC values derived from the MALDI-TOF MS method and the broth micro-dilution method was 61.67% for ceftriaxone and 71.67% for imipenem in the 60 K. pneumoniae strains (shown in Table 3).
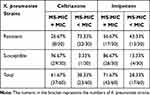 |
Table 3 The Agreement of the MIC Values Determined by MALDI-TOF MS and Broth Micro-Dilution Method |
Discussion
Similar to a previous study, the genotypic characterization of CRKP revealed that antibiotic resistance is mainly mediated by KPC-type genes and the coexpression of ESBLs and AmpC genes.27 The resistance of one CRKP strain that did not express any of the carbapenemase genes could be attributable to the ESBL and AmpC genes along with over-expression of the efflux pump or loss of outer membrane protein.28 The coexistence of resistance genes reflects the high frequency of multidrug-resistant K. pneumoniae in carbapenems, cephalosporin and aminoglycosides. Consequently, the early identification of CRKP is particularly important in the management of infectious diseases with antibiotic treatments.
In this study, we demonstrated the ability of MALDI-TOF MS to detect the growth of K. pneumoniae in a short time-frame. To allow comparison to a reference method, the inoculation volume and the turbidity of bacterial cells were standardized with the broth micro-dilution method. A recent study showed that even minimal alterations in inoculum turbidity may have a dramatic influence on the determination of MIC.29 Whilst higher bacterial cell concentrations would lead to shorter incubation time, it also increases the risk of false MS-MIC values. Compared to the broth micro-dilution method, the main difference was the separation of bacterial cells and reduced incubation time. Previous studies have demonstrated that a minimum incubation time of 4 h is optimum for the reliable detection of bacterial growth.18,23 To simplify the method, we directly centrifuged 96-well plates at 4000 rpm for 10 minutes to subside the bacterial cells rather than remove the suspension into the new 1.5 mL tube following centrifugation for 3 minutes at 12,000 rpm. The same parallel test was performed in which the suspension was pipetted into a tube and centrifuged at 12,000rpm for 3 minutes. Our results suggested that centrifugation at 4000 rpm is appropriate for MS analysis and does not negatively influence the agreement between MS-MIC and BMD-MIC.
A previous study performed on the Acinetobacter baumannii isolates showed a similar agreement of 72% in the MICs between the BMD and MS susceptibility tests of meropenem.23 The MIC values derived from MALDI-TOF MS were lower than the BMD-MIC and could be attributed to the growth characteristics of bacteria. The bacterial growth cycle is around 30 minutes and is comprised of the lag, logarithmic, stationary, and decline phases. The lag phase indicates the process of adaptation of bacteria following exposure to a new environment and its length varies with the inoculation volume of bacterial cells and antibiotic concentration.30,31 The concordance of the MIC values for ceftriaxone and imipenem between the MS and BMD methods was higher in the sensitive strains than in the resistant strains. Studies have highlighted the phenomenon of the delayed expression of resistance, indicating that some resistant strains show susceptible behaviors at early time points of incubation with antibiotics but start to grow after prolonged incubation.32,33 The disagreement in the lower MS-MIC values may reflect that the incubation time is not sufficiently long for the growth of bacterial species in the lag phase and so the number of bacteria may not be sufficient for MALDI-TOF MS detection.
The MIC values of ceftriaxone and imipenem determined by the automated micro-dilution method (VITEK-2 Compact system) were all ≥64 μg/mL and ≥16 μg/mL, respectively, in the resistant strains, and ≤1μg/mL in the susceptible isolates. The drug susceptibility card of the VITEK-2 Compact system covers serial concentrations of antibiotics that were 32, 8, 2 and 1 μg/mL for ceftriaxone, and 16, 4 and 2 μg/mL for imipenem. When compared to the MIC values determined by the VITEK-2 Compact system, the MS-based method had the same MIC results in the resistant and susceptible strains of K. pneumoniae. These data suggested that the MS susceptibility tests could optimize the concentrations range on the 96-well plate similar to the AST card of the VITEK-2 Compact system. A study performed the DOT-MGA assay at the breakpoint concentration of 2 μg/mL for meropenem to distinguish between carbapenem-susceptible and non-susceptible in 24 K. pneumoniae strains. This study acquired 100% sensitivity and 100% specificity after 4 h incubation.21 Another study used MBT-ASTRA to differentiate resistant and susceptible K. pneumoniae after 1 h of incubation at a meropenem concentration of 8 μg/mL and achieved a sensitivity of 97.3% and a specificity of 93.5%.20 The breakpoints for ceftriaxone and imipenem are both ≥4 μg/mL for resistant strains and ≤1 μg/mL for susceptible strains according to CLSI, our study suggested that MALDI-TOF MS could accurately distinguish the 30 carbapenems-resistant strains from the 30 sensitive isolates with 100% specificity and 100% sensitivity when referring to the breakpoint concentrations. However, this study did not collect the imipenem- and ceftriaxone-intermediate strains of K. pneumoniae or resistant strains that showed lower BMD-MIC values of 4–32 μg/mL. Those strains may be falsely classified as sensitive strains due to the possibility of lower MICs detected by MALDI-TOF MS.
The AST differentiates between resistant and susceptible strains independent of bacterial species, antimicrobial agents, or underlying resistance mechanisms. Our study only assessed this method for imipenem and ceftriaxone MIC detection in K. pneumoniae. The application of this approach to universal phenotypical AST requires further validation using different combinations of antibiotics with bacterial species. Further investigations are required to explore the optimum incubation time for different microbial species and to confirm the feasibility of resistance detection for different antibiotics. These studies could include further optimization of the assay, including the precoating of the 96-well plate with different antibiotics, using automatic sampling of the bacterial suspension to simplify handling and to avoid bias during manual operation, and programming a new module within the existing software to provide the final results for clinical decision-making.
Conclusion
MALDI-TOF MS is a technology that can be used for the routine identification of pathogens in the clinical laboratory and has a high value in characterizing patterns of antimicrobial resistance. This approach can be used to identify drug resistance by enzymatic hydrolysis analysis and to provide the exact MIC values of different species for antibiotics. Our method suggested that MALDI-TOF MS can be used as an alternative for the early detection of antibiotic resistance to ceftriaxone and imipenem in K. pneumoniae.
Ethical Approval
There is no ethical concern in this study, and this experiment was approved by The Medical Ethics Committee of the First Affiliated Hospital of Anhui Medical University. The written informed consent was obtained from patients in accordance with the Declaration of Helsinki.
Acknowledgment
This work was financially supported by Anhui Natural Science Foundation (grant number: 9021138201).
Disclosure
The authors declared that there are no conflicts of interest in the experimental design, data collection, manuscript drafting and revising.
References
1. Bizzini A, Greub G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect. 2010;16(11):1614–1619. doi:10.1111/j.1469-0691.2010.03311.x
2. Garza-González E, Camacho-Ortíz A, Rodríguez-Noriega E, et al. Comparison of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) and the Vitek 2 system for routine identification of clinically relevant bacteria and yeast. Ann Clin Lab Sci. 2020;50(1):119–127.
3. Tsuchida S, Umemura H, Nakayama T. Current status of matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) in clinical diagnostic microbiology. Molecules. 2020;25(20):4775. doi:10.3390/molecules25204775
4. Zhou M, Yang Q, Kudinha T, et al. An improved in-house MALDI-TOF MS protocol for direct cost-effective identification of pathogens from blood cultures. Front Microbiol. 2017;8. doi:10.3389/fmicb.2017.01824
5. Íñigo M, Coello A, Fernández-Rivas G, et al. Direct identification of urinary tract pathogens from urine samples, combining urine screening methods and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2016;54(4):988–993. doi:10.1128/JCM.02832-15
6. Florio W, Morici P, Ghelardi E, Barnini S, Lupetti A. Recent advances in the microbiological diagnosis of bloodstream infections. Crit Rev Microbiol. 2018;44(3):351–370. doi:10.1080/1040841X.2017.1407745
7. Hoyos-Mallecot Y, Riazzo C, Miranda-Casas C, Rojo-Martín MD, Gutiérrez-Fernández J, Navarro-Marí JM. Rapid detection and identification of strains carrying carbapenemases directly from positive blood cultures using MALDI-TOF MS. J Microbiol Methods. 2014;105:98–101. doi:10.1016/j.mimet.2014.07.016
8. Oviaño M, Fernández B, Fernández A, Barba MJ, Mouriño C, Bou G. Rapid detection of enterobacteriaceae producing extended spectrum beta-lactamases directly from positive blood cultures by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Clin Microbiol Infect. 2014;20(11):1146–1157. doi:10.1111/1469-0691.12729
9. Gaibani P, Galea A, Fagioni M, Ambretti S, Sambri V, Landini MP. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of KPC-producing Klebsiella pneumoniae. J Clin Microbiol. 2016;54(10):2609–2613. doi:10.1128/JCM.01242-16
10. Li J, Hu W, Li M, Deng S, Huang Q, Lu W. Evaluation of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for identifying VIM- and SPM-type metallo-β-lactamase-producing Pseudomonas aeruginosa clinical isolates. Infect Drug Resist. 2019;12:2781–2788. doi:10.2147/IDR.S211984
11. Hu Y, Cai J, Zhou H, Zhang R, Chen G. Rapid detection of porins by matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Front Microbiol. 2015;6. doi:10.3389/fmicb.2015.00784
12. Hrabák J, Chudáčková E, Walková R. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev. 2013;26(1):103–114. doi:10.1128/CMR.00058-12
13. Oviaño M, Bou G. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for the rapid detection of antimicrobial resistance mechanisms and beyond. Clin Microbiol Rev. 2019;32(1). doi:10.1128/CMR.00037-18
14. Oviaño M, Ramírez CDLL, Barbeyto LP, Bou G. Rapid direct detection of carbapenemase-producing Enterobacteriaceae in clinical urine samples by MALDI-TOF MS analysis. J Antimicrob Chemother. 2017;w579. doi:10.1093/jac/dkw579
15. Sakarikou C, Ciotti M, Dolfa C, Angeletti S, Favalli C. Rapid detection of carbapenemase-producing Klebsiella pneumoniae strains derived from blood cultures by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Bmc Microbiol. 2017;17(1). doi:10.1186/s12866-017-0952-3
16. Oviaño M, Gómara M, Barba MJ, Revillo MJ, Barbeyto LP, Bou G. Towards the early detection of β-lactamase-producing Enterobacteriaceae by MALDI-TOF MS analysis. J Antimicrob Chemother. 2017;72(8):2259–2262. doi:10.1093/jac/dkx127
17. Schuster D, Josten M, Janssen K, et al. Detection of methicillin-resistant coagulase-negative staphylococci harboring the class A mec complex by MALDI-TOF mass spectrometry. Int J Med Microbiol. 2018;308(5):522–526. doi:10.1016/j.ijmm.2018.05.001
18. Justesen US, Acar Z, Sydenham TV, Johansson Å. Antimicrobial susceptibility testing of Bacteroides fragilis using the MALDI Biotyper antibiotic susceptibility test rapid assay (MBT-ASTRA). Anaerobe. 2018;54:236–239. doi:10.1016/j.anaerobe.2018.02.007
19. Sparbier K, Schubert S, Kostrzewa M. MBT-ASTRA: a suitable tool for fast antibiotic susceptibility testing? Methods. 2016;104:48–54. doi:10.1016/j.ymeth.2016.01.008
20. Lange C, Schubert S, Jung J, Kostrzewa M, Sparbier K. Quantitative matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid resistance detection. J Clin Microbiol. 2014;52(12):4155–4162. doi:10.1128/JCM.01872-14
21. Idelevich EA, Sparbier K, Kostrzewa M, Becker K. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin Microbiol Infect. 2018;24(7):738–743. doi:10.1016/j.cmi.2017.10.016
22. Correa-Martínez CL, Idelevich EA, Sparbier K, Kostrzewa M, Becker K. Rapid detection of extended-spectrum β-lactamases (ESBL) and AmpC β-lactamases in enterobacterales: development of a screening panel using the MALDI-TOF MS-based direct-on-target microdroplet growth assay. Front Microbiol. 2019;10. doi:10.3389/fmicb.2019.00013
23. Li M, Liu M, Song Q, et al. Rapid antimicrobial susceptibility testing by matrix-assisted laser desorption ionization–time of flight mass spectrometry using a qualitative method in Acinetobacter baumannii complex. J Microbiol Methods. 2018;153:60–65. doi:10.1016/j.mimet.2018.09.002
24. Kazemian H, Heidari H, Ghanavati R, et al. Phenotypic and genotypic characterization of ESBL-, AmpC-, and carbapenemase-producing Klebsiella pneumoniae and Escherichia coli isolates. Med Princ Pract. 2019;28(6):547–551. doi:10.1159/000500311
25. Messaoudi A, Mansour W, Jaidane N, Chaouch C, Boujaâfar N, Bouallègue O. Epidemiology of resistance and phenotypic characterization of carbapenem resistance mechanisms in Klebsiella pneumoniae isolates at Sahloul University Hospital-Sousse, Tunisia. Afr Health Sci. 2019;19(2):2008–2020. doi:10.4314/ahs.v19i2.24
26. Wang G, Song G, Xu Y. Association of CRISPR/Cas system with the drug resistance in Klebsiella pneumoniae. Infect Drug Resist. 2020;13:1929–1935. doi:10.2147/IDR.S253380
27. Unlu O, Demirci M. Detection of carbapenem-resistant Klebsiella pneumoniae strains harboring carbapenemase, beta-lactamase and quinolone resistance genes in intensive care unit patients. GMS Hyg Infect Control. 2020;15:c31. doi:10.3205/dgkh000366
28. Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm. Trends Mol Med. 2012;18(5):263–272. doi:10.1016/j.molmed.2012.03.003
29. Smith KP, Kirby JE. The inoculum effect in the era of multidrug resistance: minor differences in inoculum have dramatic effect on MIC determination. Antimicrob Agents Chemother. 2018;62(8). doi:10.1128/AAC.00433-18
30. Weibull E, Antypas H, Kjäll P, Brauner A, Andersson-Svahn H, Richter-Dahlfors A. Bacterial nanoscale cultures for phenotypic multiplexed antibiotic susceptibility testing. J Clin Microbiol. 2014;52(9):3310–3317. doi:10.1128/JCM.01161-14
31. Doern CD, Forbes BA. The slow march toward rapid phenotypic antimicrobial susceptibility testing: are we there yet? J Clin Microbiol. 2018;56(4):e1917–e1999. doi:10.1128/JCM.01999-17
32. Idelevich EA, Becker K. How to accelerate antimicrobial susceptibility testing. Clin Microbiol Infect. 2019;25(11):1347–1355. doi:10.1016/j.cmi.2019.04.025
33. Adams-Sapper S, Nolen S, Donzelli GF, et al. Rapid induction of high-level carbapenem resistance in heteroresistant KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2015;59(6):3281–3289. doi:10.1128/AAC.05100-14
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

