Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 14
A Pilot Study: Composition and Diversity of 16S rRNA Based Skin Bacterial Microbiome in Indonesian Atopic Dermatitis Population
Authors Suwarsa O , Hazari MN, Dharmadji HP, Dwiyana RF , Effendi RMRA , Hidayah RMN , Avriyanti E , Gunawan H , Sutedja E
Received 9 September 2021
Accepted for publication 4 November 2021
Published 19 November 2021 Volume 2021:14 Pages 1737—1744
DOI https://doi.org/10.2147/CCID.S338550
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Oki Suwarsa, Maryam Nissa Hazari, Hartati Purbo Dharmadji, Reiva Farah Dwiyana, Raden Mohamad Rendy Ariezal Effendi, Risa Miliawati Nurul Hidayah, Erda Avriyanti, Hendra Gunawan, Endang Sutedja
Department of Dermatology and Venereology, Faculty of Medicine, Universitas Padjadjaran - Dr. Hasan Sadikin General Hospital, Bandung, West Java, Indonesia
Correspondence: Hendra Gunawan
Department of Dermatology and Venereology, Faculty of Medicine, Universitas Padjadjaran - Hasan Sadikin General Hospital, Pasteur 38, Bandung, West Java, 40161, Indonesia
Tel +62 222032426 ext. 3449
Fax +62 222032426
Email [email protected]
Background: Atopic dermatitis (AD) interferes with quality of life and is influenced by important factors like skin microbiome. The results of the skin microbiome composition and diversity in AD varied in some studies.
Purpose: This study aims to determine the composition and diversity of the skin microbiome in Indonesian AD patients.
Patients and Methods: Genomic deoxyribonucleic acid (DNA) preparations were obtained from skin swabs of the cubital fossa of 16 subjects, nine of which were having mild AD, three moderate AD, and four healthy individuals. DNA extraction and sequencing of the 16S ribosomal ribonucleic acid (rRNA) gene using next-generation sequencing and bioinformatics analysis were further performed.
Results: Firmicutes (p), Bacilli (c), Bacillales (o), Staphylococcaceae (f), and Staphylococcus (g) were dominant in moderate AD. On the contrary, Proteobacteria (p), Gammaproteobacteria (c), Pseudomonadales (o), Moraxellaceae (f), and Acinetobacter (g) were dominant in mild AD. Staphylococcus aureus was found in the highest number in individuals with moderate AD. Interestingly, Ensifer adhaerens was found in mild AD. Microbial diversity was decreased in moderate AD.
Conclusion: Metagenomic analysis in this study identified microbes in moderate and mild AD and showed a low diversity of skin microbiomes in moderate AD. Interestingly, this is the first time that the bacteria Ensifer adhaerens was detected on the human skin.
Keywords: atopic dermatitis, microbiome, skin microbiome
Introduction
Atopic dermatitis (AD) is the most common chronic, and recurrent inflammatory skin disease. The prevalence of AD varies throughout the world and still increasing. In developing countries, 10–20% of children and 1–3% of adults are affected by AD. This disease is characterized by significant itching.1,2 Intense itching causes trauma on the skin due to scratching and disturbed sleep patterns. Individuals with AD lesions can also face social stigma due to the visibility of the lesions. Additionally, chronic conditions causes individuals with AD to require ongoing treatment, in the form of daily moisturizers, repeated topical and oral therapy, and repeated consultations with doctors. These may affect the individuals, and to some extent, their family, financially.2,3
Atopic dermatitis is caused by a complex interaction between impaired skin barrier function, dysregulation of immunity, and skin microbiome.2,4 Impaired skin barrier function and immune dysregulation affect skin microbiome, which leads to an increase in Staphylococcus aureus (S. aureus) colonization.5 This condition could decrease bacterial diversity on the skin due to decreased colonization of other bacteria such as Streptococcus, Propionibacterium, Acinetobacter, Corynebacterium, and Granulicatella.5–7 The change in the microbial composition that differs from normal conditions is called skin microbiome dysbiosis.6,7
A study by Kong et al revealed a decrease of microbial diversity in skin lesions of acute exacerbation of AD due to an excessive increase in S. aureus.5 One study also reported an increase in S. aureus colonization in individuals with AD lesions that had not been treated with moisturizer compared to healthy individuals.7 However, the most predominant microbes in individuals with AD Stenotrophomonas maltophilia, which differs from that of healthy individuals.8 Kim et al found that Staphylococcus, Pseudomonas, and Streptococcus are dominant in AD skin.9
Identifying specific bacteria in individuals with AD is difficult. Therefore, it is necessary to conduct further microbial profile studies in individual AD populations. Molecular techniques help detect various microbes that have not yet been identified in human organs.8 Deoxyribonucleic acid (DNA) sequencing method against 16S ribosomal ribonucleic acid gene (16S rRNA gene) in bacteria produces a more outstanding picture of skin microbiome diversity compared to the culture method,6 as this method can detect bacteria that are difficult to culture or isolate.8 This study aims to determine the composition and diversity of the skin microbiome in Indonesian AD patients.
Materials and Methods
Study Population and Inclusion Criteria
The study was approved by the Institutional Review Board of Health Research Ethics Committee in Dr. Hasan Sadikin Hospital, Bandung, West Java, Indonesia and conducted in accordance with the latest version of the Declaration of Helsinki. Each subject and their parents gave written informed consent and consent to being photographed prior to enrolment.
The subjects were 12 AD patients that had an onset or exacerbation and four normal individuals. Dermatologists clinically diagnosed the AD patients, then evaluated the severity of their condition using the Scoring Atopic Dermatitis (SCORAD) index, a complex measures that assess a weighted average of the severity of AD signs and extent of lesions.10 The inclusion criteria were AD patients and their parents gave their consent to participate in the study, The exclusion criteria were subjects who had another skin disease, received topical or oral antibiotics or corticosteroids within the previous seven days, applied emollient routinely within the last seven days, or have bathed within the last 12 hours. For normal individuals agree to participate in the study and the exclusion criteria were have previous skin diseases, including AD, allergic or significant underlying diseases, and history of taking medications, including corticosteroid, antibiotic, and emollient.
Data Collection (Sampling/Collection of Skin Swab)
A cross-sectional descriptive study with consecutive sampling was performed from January 13 to July 13, 2020. For all subjects, the following data were collected: sex, age, education, occupation, AD duration, the onset of AD symptoms, medication history, and disease severity as assessed using the SCORAD index. Microbial skin samples were collected using skin swab taken with single-use sterile cotton-tipped swabs. Swabs were pre-moistened with a sterile buffer solution and rubbed firmly for 20 seconds over affected skin at the volar region of the forearm (cubital fossa). The cotton-tip samples were then stored in a tube filled with a sterile buffer solution and shipped to the Genetika Science Laboratory for processing.
DNA Extraction, Polymerase Chain Reaction (PCR) Amplification, Next Generation Sequencing (NGS), and NGS Analyses
Genomic DNA was extracted from each swab using the Qiagen Gel Extraction Kit (Qiagen, Germany). The 16S rRNA genes of distinct regions (16SV3-V4) were amplified using a specific primer of variable regions 3 through 4. Primers 341F: CCTAYGGGRBGCASCAG and 806R: GGACTACNNGGGTATCTAAT were used for the amplification of regions. The following PCR conditions were: 95℃ for 1 minute followed by 35 cycles of 95℃ for 15 seconds, 52℃ for 15 seconds, and 72℃ for 10 seconds,Paired-end reads were assigned to samples based on their unique barcodes and truncated by cutting off the barcode and primer sequences. Paired-end reads were merged using FLASH, a fast and accurate analysis tool designed to merge paired-end reads when at least some reads overlap the read generated from the opposite end of the same DNA fragment.11 The splicing sequences were called raw tags. Quality filtering on the raw tags was performed under specific conditions to obtain the high-quality clean tags according to the QIIME quality-controlled process.12,13 The tags were then compared with the reference database using UCHIME algorithm to detect chimera sequences. The chimera sequences were removed, and the Effective Tags were finally obtained.14,15
Sequence analysis was performed by Uparse software using all the effective tags. Sequences with ≥97% similarity were assigned to the same Operational Taxonomic Unit (OTU).16 A representative sequence for each OTU was screened for further annotation. For each representative sequence, Mothur software was used to compare against the small subunit ribosomal ribonucleic acid (SSU rRNA) database of SILVA Database for species annotation at each taxonomic rank (Threshold: 0.8~1) (kingdom, phylum, class, order, family, genus, species).17,18 To obtain the phylogenetic relationship of all OTUs representative sequences, the MUSCLE (Version 3.8.31) was used to compare multiple sequences rapidly.19 OTUs abundance information was normalized using a standard sequence number corresponding to the sample with the least sequences.
Alpha diversity is applied in analyzing the complexity of biodiversity for a sample through 6 indices, including Shannon and Simpson. All the indices in our samples were calculated with QIIME (Version 1.7.0) and displayed with R software (Version 2.15.3). Subsequent analysis of alpha diversity was all performed based on this output normalized data.
Results
Demographics of Study Participants
The majority of the study participants with AD were women (58.3%), aged >18 years (58.3%), with a bachelor degree (75%), and were housewives (33.4%). All subjects had <6 months duration of AD (100%), with most of them having their AD exacerbation at >18 years and starting in early life (50%). Mild AD was the most frequently found, based on the SCORAD assessment (75%). All participants in the current study had a history of previous treatment (Table 1). While the normal group consisted of 2 females and 2 males, it consisted of 1 participant <10 years old and 3 other participants over 18 years old. These four normal subjects’ occupations and education were a kindergarten pupil, a gardener with high school graduated, a medical doctor, and a houseworker high school graduated.
 |
Table 1 Demographics of Study Participants with Atopic Dermatitis |
Overview of Skin Microbiomes in Study Participants with Atopic Dermatitis
Skin microbiome composition in patients with AD was investigated using 16S rRNA next-generation sequencing. The most abundant phylum in moderate AD was the Firmicutes phylum, followed by the Proteobacteria and Actinobacteria phyla (Figure 1A). In contrast, the skin microbiome of mild AD patients and healthy individuals were predominated by Proteobacteria, followed by Firmicutes and Actinobacteria (Figure 1A). At the class level, the predominant bacteria in moderate AD were of the Bacilli class, was followed by Alphaproteobacteria and Gammaproteobacteria. Gammaproteobacteria were more frequent in mild AD, followed by Alphaproteobacteria and Bacilli. Furthermore, Bacilli, Unidentified Actinobacteria, and Gammaproteobacteria were dominant in healthy individuals as shown in Figure 1B.
 |
Figure 1 The most abundant phylum (A) classes (B) order (C) family (D) and genus (E) on the skin of AD patients. |
The most abundant order in moderate AD were Bacillales, followed by Rhizobiales and Micrococcales. Moreover, mild AD was predominated by Pseudomonadales, followed by Bacillales, Micrococcales, and Rhizobiales. The most predominant order in healthy individuals was Bacilles, followed by Micrococcales and Pseudomonadales, as shown in Figure 1C. Furthermore, Staphylococcaceae, Rhizobiaceae, and Bacillaceae were the dominant family in moderate AD. In contrast, Moraxellaceae, Rhodobacteraceae, Staphylococcaceae, and Rhizobiaceae made up the most prominent family in mild AD, whereas Staphylococcaceae, Intrasporangiaceae, and Moraxellaceae were abundant in healthy individuals (Figure 1D). The most observed genus in moderate AD was Staphylococcus, followed by Ensifer and Bacillus. Furthermore, Acinetobacter was the most abundant genus in mild AD, followed by Paracoccus, Staphylococcus, and Ensifer. In contrast, Staphylococcus, Knoellia, and Acinetobacter were dominant in healthy individuals (Figure 1E).
In mild AD, Ensifer adhaerens (12.8%) accounted for the highest proportion of species, followed by S. aureus (10.17%) and Pseudomonas stutzeri (6.53%). On the other hand, in moderate AD, the most prominent species found was S. aureus (49.32%), followed by Ensifer adhaerens (29.8%) and B. aerophilus (9.55%). Bacterial diversity was also analyzed using 16S rRNA next-generation sequencing, and the highest diversity of skin microbiomes was seen in mild AD. Moderate AD patients had lower microbial diversity as measured by Shannon and Simpson indices, as shown in Figure 2.
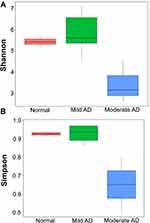 |
Figure 2 The skin microbiome diversity as assessed using the Shannon index (A) and Simpson index (B). |
Discussion
The skin is the most exposed organ of the body to environmental changes and stress. It has a dynamic and complex microbial ecosystem. The bacteria that live on the skin are closely related in type and density to those found in skin glands or hair follicles in different areas. For example, Staphylococcus is dominant in the sebaceous and the moist regions, while Propionibacterium dominates only in the sebaceous and Corynebacterium only in the moist areas.20 A complex microbial community termed “the microbiome” colonizes the skin.21 There are links between intestinal microbiota and autoimmune diseases (arthritis, psoriasis, diabetes, and others) that target different tissues (joints, skin, and others), not only those that target the intestine. The most common immune-mediated skin diseases are atopic dermatitis, psoriasis, vitiligo, scleroderma, pemphigus vulgaris, bullous pemphigoid, lichen planus, erythema multiforme, and systemic lupus erythematosus (SLE).20
Fahlen et al showed that the level of Actinobacteria was significantly increased in the skin from healthy individuals compared with that of psoriasis patients, while Proteobacteria dominated in lesional skin.22 Additionally, they observed a higher ratio of Streptococcus/Proteobacteria in the skin of these patients. A study reported by Ganju et al analyzed the cutaneous microbiota in patients with vitiligo. They observed a reduction of microbiota diversity in lesional sites compared with non-lesional sites; Actinobacteria represented the dominant taxa in non-lesional skin while Firmicutes and Proteobacteria dominated in the lesional areas.23 Only a few studies have shown the presence of dysbiosis in patients with SLE. Specifically, a lower ratio of Firmicutes/Bacteroides was identified in SLE patients in Spain and southern China. The increase in Actinobacteria was only significant in patients from the latter region.20
Metagenomic analysis using 16S rRNA next-generation sequencing can detect bacteria in greater numbers and in more variations compared to conventional methods such as bacterial culture.6,8 This culture-independent method makes it possible to identify and count the relative abundance of all species present. The results of this study support the results of previous studies for moderate AD, although the results also vary in several studies. The skin microbiome composition found in moderate AD in this study was similar to those of Kim et al and Flores et al with moderate AD participants. Kim et al reported the most genera detected in patients with moderate AD in Korea was Staphylococcus.9 Other genera found in abundance include Pseudomonas, Streptococcus, and Acinetobacter. Lactobacillus, Xanthomonadaceae, Leuconostoc, Alcaligenaceae, Lactococcus, Rhizobiales, and Methylobacterium, although their composition was relatively small. In addition to genus level, most of the bacteria detected in AD patients were of the Staphylococcaceae, Pseudomonadaceae, and Streptococcaceae families. At the order level, the majority of bacteria came from the Bacillales and Pseudomonadales orders, while at the class level, most of the bacteria came from the Bacilli class. At the phylum level, the majority of bacteria were from the Firmicutes phylum. In healthy individuals, the most dominant bacteria detected at the family level were Alcaligenaceae, followed by Sediminibacterium and Lactococcus; at the order level, Burkholderiales was found to be most abundant; while at the class level it was Betaproteobacteria and at the phylum level it was Proteobacteria.9
A study of skin microbiomes from the United States by Flores et al on moderate AD patients before and after using moisturizers showed that Staphylococcus was the most frequent genus on the skin of patients with AD before moisturizing. Other genera found in patients with AD were Propionibacterium, Streptococcus, Alicyclobacillus, Corynebacterium, Stenotrophomonas, Streptococcaceae, Acinetobacter, and Prevotella.7
The composition of the skin microbiome of healthy individuals in this study was similar to the results of several studies. Gao et al from the United States reported the molecular analysis of healthy individual skin bacteria from the volar surface of the forearm.24 This study showed that there were three phyla found, Actinobacteria, Firmicutes, and Proteobacteria. There were four genera detected, namely Propionibacteria, Corynebacteria, Staphylococcus, and Streptococcus.24
One study from the United States reported that the sequencing results of five samples taken from the volar forearm of healthy individuals with no history of skin disease were the Proteobacteria group with Pseudomonas being the most frequent genus, followed by Janthinobacterium.25 Other identified genera were Serratia, Halomonas, Stenotrophomonas, Delftia, and Comamonas. The Actinobacteria group consisted of the genera Corynebacterium, Kocuria, Propionibacterium, Microbacterium, and Micrococcus. Firmicutes were the third most-frequently identified, with the genus Staphylococcus and Clostridium being the most abundant. Furthermore, the fourth phylum detected was Bacteroidetes, with the predominant genus being Sphingobacterium or Chryseobacterium.25 Another study analyzed the skin microbiota in healthy individuals from 10 study participants. The study detected 16 phyla with the most dominant phyla being Actinobacteria (51.8%), Firmicutes (24.4%), Proteobacteria (16.5%), and Bacteroidetes (6.3%). Of the 12 genera detected, the most dominant ones were Corynebacteria (22.8%), Propionibacteria (23.0%), and Staphylococci (16.8%).26 Washing hands might increase the Staphylococcaceae, Streptococcaceae, and Lactobacillaceae on the skin of the hands. However, it did not change the composition of the majority of bacteria on the skin.27
In our study, the highest number of species detected in individuals with moderate AD was S. aureus. It was similar to Kim et al’s study that found S. aureus as the most frequently found species in AD patients, followed by S. epidermidis and S. haemolyticus.9 Flores et al also reported that S. epidermidis, S. aureus, and S. haemolyticus as the most abundant bacteria found in AD patients.7 Dekio et al reported that the skin microbiota in AD patients mostly consists of Staphylococcus sp. This study also discovered a new species that have never been reported before in the skin of AD patients. The new species or phylotype detected in patients with AD was Stenotrophomonas maltophilia. While D. maris, also a never-before-seen species, was found in normal individuals.8 The burden of S. aureus on lesional and non-lesional skin correlates with the severity of the disease. S. aureus directly influences pathogenesis, and several factors produced by the bacterium increase inflammation and exacerbate AD symptoms, including staphylococcal superantigen B and delta-toxin.28
From 102 AD patients in Kong et al’s study, the most detected bacteria came from the genus Staphylococcus, including S. epidermidis and S. aureus, followed by bacteria from the Streptococcus, Corynebacterium, and Propionibacterium genera.5 Grice et al reported that Propionibacteria and Staphylococci were the most numerous species in the sebaceous regions. In the moist skin areas, such as nostrils, armpits, volar arms, between fingers, hips, buttocks, behind the knees, and umbilicus, most species found were Corynebacteria, although Staphylococci also been identified.26
Flores et al reported that the diversity of microbiome in AD lesions was reduced and, there was a significant decrease in the genus Staphylococcus and a significant increase in the variety of skin microbiome (p = 0.01) post-administration of moisturizers.7 Other study using dupilumab as treatment significantly changed the skin microbiome and decreased S. aureus.29 We also studied the skin microbiomes of mild AD patients. In this study, the highest diversity of skin microbiome was seen in samples of individuals with mild AD, and the lowest diversity was seen in individuals with moderate AD. Kong et al found that treatment changed the bacterial skin diversity in AD and suggested that treatments diversify skin bacteria preceding improvements in disease activity. Dramatic reductions in the skin microbial diversity occurred during AD flares, restored with standard AD therapies. In AD, the proportion of Staphylococcus sequences, particularly S. aureus, was more significant during disease flares than at baseline or post-treatment and correlated with worsened disease severity.5 Higher S. aureus abundances in patients with AD relative to healthy subjects, in lesional sites, compared to non-lesional sites, and in patients with higher vs lower SCORAD also revealed in Khadka et al’s study.30
Furthermore, the most abundant species found in mild AD was Ensifer adhaerens, a bacterial species of the genus Ensifer. The bacteria was first isolated in 1982 and 1983 and located in the soil.31 No study had previously detected this bacteria on the human skin. The microbiota is generally conceived of as two groups, resident and transient microbes.5 In this study, Ensifer adhaerens could be as transient microbes that do not establish themselves permanently on the skin’s surface but rather arise from the environment and persist for hours to days. Furthermore, people with soil-related occupation or daily activities could be the source of inoculation of the soil bacteria, and soil is the habitat of the Ensifer adhaerens.31 Additionally, Indonesia has high humidity conditions, about 43–98%, making the bacteria grow high.32
Conclusion
Metagenomic analysis in this study succeeded in identifying microbes in moderate and mild AD and showed a low diversity of skin microbiome in moderate AD. The Ensifer adhaerens detected in mild AD is a newly detected bacteria on human skin.
Acknowledgments
The Author would like to thank all of the staff Dermatology and Venereology Department, Faculty of Medicine, Universitas Padjadjaran – Dr. Hasan Sadikin General Hospital.
Funding
This study was supported by Academic Leadership Grant from Universitas Padjadjaran, Bandung, West Java, Indonesia.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Simpson EL, Leung DYM, Eichenfield LF, et al. Atopic dermatitis. In: Kang S, Amagai M, Bruckner AL, Enk AH, Margolis DJ, McMichael AJ, editors. Fitzpatrick’s Dermatology.
2. Kapur S, Watson W, Carr S. Atopic dermatitis. Allergy Asthma Clin Immunol. 2018;14(2):43–52. doi:10.1186/s13223-018-0281-6
3. Barbeau M, Bpharm HL. Burden of atopic dermatitis in Canada. Int J Dermatol. 2006;45(1):31–36. doi:10.1111/j.1365-4632.2004.02345.x
4. Kim JK, Kim BE, Leung DYM. Pathophysiology of atopic dermatitis: clinical implications. Allergy Asthma Proc. 2019;40(2):84–92. doi:10.2500/aap.2019.40.4202
5. Kong HH, Oh J, Deming C. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. doi:10.1101/gr.131029.111
6. Williams MR, Gallo RL. The role of the skin microbiome in atopic dermatitis. Curr Allergy Asthma Rep. 2015;15(11):65. doi:10.1007/s11882-015-0567-4
7. Flores GE, Seité S, Henley JB, et al. Microbiome of affected and unaffected skin of patients with atopic dermatitis before and after emollient treatment. J Drugs Dermatol. 2014;13(11):611–618.
8. Dekio I, Sakamoto M, Hayashi H. Characterization of skin microbiota in patients with atopic dermatitis and in normal subjects using 16S rRNA gene-based comprehensive analysis. J Med Microbiol. 2007;56:1675–1683.
9. Kim MH, Rho M, Choi JP, et al. A metagenomic analysis provides a culture-independent pathogen detection for atopic dermatitis. Allergy Asthma Immunol Res. 2017;9(5):453–461. doi:10.4168/aair.2017.9.5.453
10. Silverberg JI, Lei D, Yousaf M, et al. What are the best endpoints for eczema area and severity index and scoring atopic dermatitis in clinical practice? A prospective observational study. Br J Dermatol. 2021;184(5):888–895. doi:10.1111/bjd.19457
11. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinform. 2011;27(21):2957–2963. doi:10.1093/bioinformatics/btr507
12. Bokulich NA, Subramanian S, Faith JJ, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10(1):57–59. doi:10.1038/nmeth.2276
13. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi:10.1038/nmeth.f.303
14. Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinform. 2011;27(16):2194–2200. doi:10.1093/bioinformatics/btr381
15. Haas BJ, Gevers D, Earl AM, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. doi:10.1101/gr.112730.110
16. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi:10.1038/nmeth.2604
17. Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi:10.1128/AEM.00062-07
18. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596.
19. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi:10.1093/nar/gkh340
20. Catinean A, Neag MA, Mitre AO, Bocsan CI, Buzoianu AD. Microbiota and immune-mediated skin diseases—An overview. Microorganisms. 2019;7(9):279. doi:10.3390/microorganisms7090279
21. Nørreslet LB, Agner T, Clausen ML. The skin microbiome in inflammatory skin diseases. Curr Dermatol Rep. 2020;9(2):141–151. doi:10.1007/s13671-020-00297-z
22. Fahlén A, Engstrand L, Baker BS, Powles A, Fry L. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Arch Dermatol Res. 2012;304(1):15–22. doi:10.1007/s00403-011-1189-x
23. Ganju P, Nagpal S, Mohammed M, et al. Microbial community profiling shows dysbiosis in the lesional skin of Vitiligo subjects. Sci Rep. 2016;6(1):18761. doi:10.1038/srep18761
24. Gao Z, Tseng C, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104(8):2927–2932. doi:10.1073/pnas.0607077104
25. Grice EA, Kong HH, Renaud G, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18(7):1043–1050. doi:10.1101/gr.075549.107
26. Grice EA. Topographical and temporal diversity of the human skin microbiome. Science. 2009;3(24):1190–1192. doi:10.1126/science.1171700
27. Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105(17):994–999. doi:10.1073/pnas.0807920105
28. Towell AM, Feuillie C, Vitry P, et al. Staphylococcus aureus binds to the N-terminal region of corneodesmosin to adhere to the stratum corneum in atopic dermatitis. Proc Natl Acad Sci USA. 2021;118(1):e2014444118. doi:10.1073/pnas.2014444118
29. Olesen CM, Ingham AC, Thomsen SF, et al. Changes in skin and nasal microbiome and Staphylococcal species following treatment of atopic dermatitis with dupilumab. Microorganisms. 2021;9(7):1487. doi:10.3390/microorganisms9071487
30. Khadka VD, Key FM, Romo-González C, et al. The skin microbiome of patients with atopic dermatitis normalizes gradually during treatment. Front Cell Infect Microbiol. 2021:910. doi:10.3389/fcimb.2021.720674
31. Germida J, Casida LE. Ensifer adhaerens predatory activity against other bacteria in soil, as monitored by indirect phage analysis. Appl Environ Microbiol. 1983;45(4):1380–1388. doi:10.1128/aem.45.4.1380-1388.1983
32. Pratiwi C, Rahayu WP, Lioe HN, Herawati D, Broto W, Ambarwati S. The effect of temperature and relative humidity for Aspergillus flavus BIO 2237 growth and aflatoxin production on soybeans. Int Food Res J. 2015;22(1):82–87.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
