Back to Journals » Cancer Management and Research » Volume 13
A Novel Circular RNA circPTCD3 Promotes Breast Cancer Progression Through Sponging miR-198
Authors Zhang Z, Hu H, Li Q, Yi F, Liu Y
Received 30 March 2020
Accepted for publication 25 August 2020
Published 10 November 2021 Volume 2021:13 Pages 8435—8443
DOI https://doi.org/10.2147/CMAR.S256091
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Bilikere Dwarakanath
Zhaohui Zhang,1 Hao Hu,2 Qian Li,1 Fumei Yi,1 Yan’e Liu1
1Department of Tumor Chemotherapy and Radiation Sickness, Peking University Third Hospital, Beijing, 100191, People’s Republic of China; 2Beijing 100biotech Co., Ltd., Beijing, 100006, People’s Republic of China
Correspondence: Zhaohui Zhang
Department of Tumor Chemotherapy and Radiation Sickness, Peking University Third Hospital, Huayuan Bei Road, Haidian District, Beijing, 100191, People’s Republic of China
Email [email protected]
Introduction: Circular RNAs (circRNAs), a new type of non-coding RNA, have been demonstrated to play critical roles in the progression of various of malignant cancers. However, the function of circRNAs in breast cancer is not clearly understood.
Methods: qPCR was used to evaluate the gene expression. Function studies including MTT, transwell, wound healing and colony formation assay were performed to evaluate the function of circPTCD3 in breast cancer. Luciferase and RNA pull-down assays were used to verify the interaction between circPTCD3 and miR-198. The xenograft model was established to evaluate the function of circPTCD3 in vivo.
Results: In the present study, we identified a novel circRNA termed as circPTCD3 which was indicated to be significantly up-regulated in breast cancer tissues and cell lines. The results revealed that ectopic expression of circPTCD3 promoted the cell proliferation, migration and colony formation ability of breast cancer cells. Constantly, silencing of circPTCD3 inhibited those of breast cancer cells. Furthermore, we identified that circPTCD3 was able to target miR-198 in breast cancer cell. miR-198 has the function of inhibiting proliferation and migration of breast cancer cells which can be reversed by circPTCD3.
Conclusion: Taken together, our findings for the first time identified a novel circRNA (circPTCD3) and revealed its oncogenic role in breast cancer. Mechanically, we reported that circPTCD3 served as a competing endogenous RNA (ceRNA) to sponge miR-198. These findings provide insights into breast cancer progression and also potential new targets for diagnosis or treatment of breast cancer.
Keywords: circPTCD3, breast cancer, miR-198
Introduction
Breast cancer (BC) is counted among the key causes leading to cancer death across the globe, and is named as one of the most frequently found cancers in women worldwide.1 Despite considerable advances in both the therapy and diagnosis of breast cancer, no effective treatment has been developed for such a type of hormone-insensitive cancer.2,3 The currently available treatment for breast cancer is considered challenging even now. Accordingly, the development of innovative drugs for treating BC with high specificity and effect is required.
Circular RNAs (circRNAs) are an innovative class of the noncoding RNAs, characterized by a covalently closed loop without 5ʹ–3ʹ polyadenylated tails, which are developed by backsplicing events.4 Currently, circRNAs have been illustrated as highly conserved and abundant with high stability across various species.5 With numerous investigations underway, circRNA was observed as differentially expressed in various of cancers, which include prostate cancer, breast cancer and hepatocellular carcinoma.6–8 Also, they were indicated as having involvement in a number of physiological and pathophysiological processes including cell proliferation, invasion, apoptosis and differentiation.
MicroRNAs (miRNAs) are small endogenous noncoding RNAs capable of regulating the cell process by means of binding to the specific target genes.9 This binding has the potential to inhibit translation or induce the degradation of target mRNAs. CircRNAs primarily sponge miRs to exert an impact on not only the gene expression but also biological functions.10
In the current research work, we figured out an up-regulated circular RNA hsa_circ_0055478 (circPTCD3) in both breast cancer tissues and cell lines. The loss and gain of function studies were carried out for the determination of the effect of circPTCD3 on tumorigenesis of breast cancer. Mechanistically, we predicted and verified the target miRNA of circPTCD3, which is miR-198. To summarize, circ-PTCD3 works as an oncogene in breast cancer, besides being likely to constitute a potential therapeutic target for breast cancer.
Methods and Materials
Patients and Specimens
Twenty-four breast cancer tumor tissue samples and adjacent normal tissues were collected from Peking University Third Hospital. The tissues were frozen in liquid nitrogen immediately after excision and then stored in −80 °C. All experimental procedures were approved by the Ethics Committee of Peking University Third Hospital and in accordance with the Declaration of Helsinki of the World Medical Association. The written informed consent was obtained from each patient.
Cell Culture
The normal breast cell line MCF-10a and breast cancer cell lines (MCF-7, BT474, MDA-MB-231, SKBr-3) were purchased from Chinese Academy of Sciences. The culturing of cells was carried out in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS, Hyclone, USA). Each cell line was cultured at 37°C with 5% CO2. In addition, the cells were treated with the indicated concentration, whereas the control cells received an equivalent amount of DMSO. The final concentration of DMSO is below 0.1%.
Cell Transfection
miR-198 mimics and mimic control were synthesized by GenePharma (Shanghai, China). CircPTCD3 overexpressing vector (PLO-ciR/circPTCD3), siRNA for circPTCD3 (si-circPTCD3) and short hairpin RNA (shRNA) targeting circPTCD3 (sh-circPTCD3) were purchased from Ribobio (Guangzhou, China). Cells were seeded on six-well plates at the density of 8×105, followed by getting cultured in RPMI-1640 without FBS at 37°C for 12 h prior to the transfection. Cell transfection and co-transfection were carried out with the use of Lipofectamine 2000 (Invitrogen, CA, USA) in accordance with the manufacturer’s instructions. 6 h following the transfection, the cell culture medium was replaced with RPMI-1640 medium, which was supplemented with 10% FBS. Cell lines stably expressing circPTCD3 or sh-circPTCD3 were established by selecting with G418 for 6 weeks.
MTT
Twenty-four hours following the transfection, cells were collected and resuspended in culture medium, followed by seeding on 96-well plates at a density of 1×104 cells/well. Moreover, 10 µL CCK-8 solution was added to each well and incubated for another 2 h at 37°C. Absorbance at 490 nm was detected using a microplate Reader (Biorad, CA, USA). Each sample was performed for at least 3 times.
Transwell Assay
Subsequent to the transfection, 1×105 of cells suspended in 200μL RPMI-1640 medium were seeded onto the upper transwell chamber (Corning, NY, USA). Following 24 hours of incubation at 37°C, the cells on the surfaces of lower chambers were fixed using 20% methanol followed by staining with 1% crystal violet (Bicobio, Tianjin, China). Eventually, the cell colonies were both photographed and counted.
Wound Healing Assay
Subsequent to the transfection, the cells were seeded onto six-well plates followed by culturing in serum free medium for a period of 24 hours. Thereafter, cell monolayers were wounded using a 10μL pipette tip. After that, cells were replaced with fresh medium and cultured at 37°C. The pictures were taken 48 after the wounding for the purpose of determining the wound-closing procedure.
Colony Formation Assay
Following the transfection, the cells were seeded in 12-well plates at the density of 100/well. Subsequent to the incubation for 2 weeks, cells were fixed in 10% formaldehyde, together with staining with 1% crystal violet (Beyotime, Shanghai, China). Images were photographed under a microscope (Leica, Germany), besides counting the colonies that contained more than 50 cells.
Xenograft Model
All the animal experimental procedures were approved by the Ethics Committee of Peking University Third Hospital. All animal procedures were performed in accordance with institutional guidelines and approved by the Animal Studies Committee of Peking University Third Hospital. MCF-7 cells were harvested and resuspended in RPMI-1640 medium. The nude mice were injected with a total number of 3 × 106/100 μL cells subcutaneously at the posterior flank. In addition, tumour size was monitored through the measurement of the length (L) and width (W) using callipers every 3 days. Following a period of 28 days, the tumours were excised out from the sacrificed mice and weighed.
PCR
Total RNA was extracted with the use of TRIzol reagent (Invitrogen, CA, USA) in accordance with the manufacturer’s instructions. Afterwards, RNA was reverse transcribed to cDNA with the help of a PrimeScript RT Reagent kit (Takara, Dalian, China). The real-time qPCR analysis was carried out with the help of a SYBR-Green PCR Master Mix (Thermo Fisher, MA, USA) in a 7900 PCR System (ABI, CA, USA). The use of GAPDH and U6 was made as internal controls for the mRNA and miRNA analysis, correspondingly. The relative expression levels were assessed with the use of the 2−ΔΔCt methodology. All of the reactions were carried out in triplicate.
Fluorescence in situ Hybridization (FISH)
Alexa Fluor 555-labeled circPTCD3 probes were designed and synthesized by RiboBio (Guangzhou, China). FISH experiment was carried out using a fluorescent in Situ Hybridization Kit (RiboBio, Guangzhou, China). Then, 1×105 cells were seeded onto the slides and cultured for a period of 24 h. Subsequent to fixing with 4% paraformaldehyde for 20 min, followed by the permeabilization with 0.5% Triton X-100 for 10 min, the cells were cultured at 37°C overnight. Eventually, the incubation of slides was carried out with DAPI for the purpose of staining the cell nucleus and observed under a fluorescence microscope (Leica, Wetzlar, Germany).
RNA Pull Down
The biotin-labelled circPTCD3 and miR-198 probe and their control probes were synthesized by Sangon Biotech (Shanghai, China). In addition, the probe-coated beads were generated through the co-incubation of the probe with the streptavidin-coated beads (Invitrogen, CA, USA) at 25°C for 2 h. Cells were gathered and incubated with circPTCD3 probes overnight at 4°C. Following that, the beads were eluted and the complex was purified with TRIzol (Takara, Dalian, China). Afterwards, the abundance of both circPTCD3 and miR-198 was analyzed by the qPCR.
IHC
The tumour tissues were fixed in 4% paraformaldehyde for 24 h, followed by dehydrating in a graded alcohol series and embedding in paraffin. The tissues were cut into 5μm sections. The sections were deparaffinised, rehydrated with a graded alcohol series and then incubated in 96°C with 0.01 mol/L sodium citrate buffer for the antigen retrieval. Following the incubation in 5% H2O2 for a period of 2 h, the sections were incubated using anti-ki67 antibody overnight at 4°C. Immunostaining was carried out with the use of streptavidin–peroxidase and diaminobenzidine (DAB) following the manufacturer’s instructions (Beyotime, Shanghai, China). Eventually, the sections were not only observed under a fluorescence microscope (Leica, Wetzlar, Germany) but also imaged.
Luciferase Reporter Assay
Cells were co-transfected with miR-198 mimics or mimic control along with pGL3 reporter vectors (Promega, CA, USA) that contain the wild-type (Wt) or mutated (mut) binding sequence of circPTCD3. Forty-eight hours following the transfection, cells were gathered and analyzed with a Dual-Luciferase Reporter Assay kit (Promega, CA, USA). Luciferase activity was detected through the use of a GloMax fluorescence reader (Promega, CA, USA). Renilla luciferase activities were used as an internal control. Each of the assays was carried out in a minimum of triplicate.
Statistics Analysis
The results are expressed as mean ± SD. Statistical analyses were performed using SPSS 19.0 software (IBM, NY, USA). Student’s t-test or one-way ANOVA were used to assess the statistical significance. Pearson analysis was carried out to assess the correlation. P<0.05 and was considered statistically significant.
Results
The Expression of circPTCD3 Was Elevated in Breast Cancer Tissues and Cells
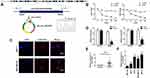
To explore the role of circPTCD3 in the progression of breast cancer, we first figured out the mechanism of ring formation. In accordance with the UCSC Genome Browser Home, circPTCD3 is produced at the PTCD3 gene locus, which contains exons 3–9. Subsequent to that, the head-to-tail splicing junction of has_circPTCD3 was verified by means of the Sanger sequencing, as presented in Figure 1A. Breast cancer cells were treated with Actinomycin D in order to inhibit the transcription after which the half-life of circPTCD3 was assessed. The results shed light on a longer half-life of circPTCD3 as compared with that of linear PTCD3 (Figure 1B). Besides that, a specific probe was designed and made to carry out the FISH experiment. Figure 1C demonstrates that circPTCD3 located primarily in cytoplasm. As we know, circRNAs have much more stability for their looped structure. Accordingly, for the purpose of confirming the stability of circPTCD3, RNase R was employed in the experiments. As the results suggested, following the RNase R treatment, the linear forms of PTCD3 showed an evident decline while no significant change was observed in the level of circPTCD3, indicating the high stability of circPTCD3 (Figure 1D). Then, we detected the expression level of circPTCD3 with the use of qPCR. The results illustrated that circPTCD3 was remarkably up-regulated in the breast cancer tissues and cells (Figure 1E and F).
CircPTCD3 Promoted the Proliferation and Migration of Breast Cancer Cells
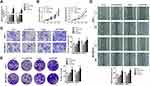
Due to the upregulated expression in breast cancer tissues and cell lines, we speculated that circPTCD3 plays quite a crucial role in the progression of breast cancer. The gain and loss of function studies were carried out with the use of overexpressing vector (PLO-ciR/circPTCD3) and siRNA for circPTCD3 (si-circPTCD3). The qPCR analysis verified the overexpression or knockdown efficiency of the PLO-ciR/circPTCD3or si-circPTCD3, correspondingly (Figure 2A). As the MTT results suggested, the circPTCD3 overexpression promoted cell proliferation while the circPTCD3 knock down inhibited that of breast cancer cells (Figure 2B). Furthermore, wound healing and transwell assay were carried out for the evaluation of the migration ability of breast cancer cells. As the results suggested, circPTCD3 overexpression reduced the migrated distance in the wound healing assay as well as the migrated cell number in transwell assay. On the contrary, the silencing of circPTCD3 lowered the capability of breast cancer cells as compared with the siRNA control group (Figure 2C and D). Colony formation was further carried out for the confirmation of our speculation. As expected, the circPTCD3 overexpression augmented the colony formation capability, while silencing of circPTCD3 lowered it in breast cancer cells (Figure 2E).
CircPTCD3 Exerts Its Function by Sponging miR-198
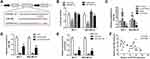
It is well known that circRNAs sponge miRNAs for regulating the downstream gene expression. We carried out the bioinformatics analysis software, CircInteractome, aimed at predicting the potential miRNA target of circPTCD3, followed by selecting miR-198. Figure 3A demonstrates the complementary base sequences between circPTCD3 and miR-198. The luciferase activity assay was carried out for the verification of the binding of circPTCD3 and miR-198. We discovered the fact that miR-198 lowered the luciferase activities of the wild-type reporter for circPTCD3 but not that of the mutant-type reporter, confirming miR-198 as a sponge target of circPTCD3 (Figure 3B). Subsequent to that, we found that circPTCD3 overexpression lowered the level of miR-198 while the circPTCD3 knock down increased that (Figure 3C). The RNA pull down was further carried out for the purpose of confirming the binding of circPTCD3 and miR-198. By means of the results, we figured it out that the enrichment of miR-198 was substantially higher in the circPTCD3 probe pull-down product in comparison with that of the control probe (Figure 3D). Accordingly, more circPTCD3 was captured by the biotin-labelled miR-198 groups as compared with the control probe (Figure 3E). Eventually, the Pearson analysis was carried out to determine the relationship between circPTCD3 and miR-198. As the results suggested, there was a significant negative relationship existing between circPTCD3 and miR-198 (Figure 3F). These findings suggested miR-198 as a sponge target of circPTCD3 in breast cancer cells.
CircPTCD3 Reversed the Function of miR-198 in Breast Cancer Cells
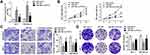
Being a target of circPTCD3, we predicted that miR-198 had involvement in the biological mechanism of breast cancer cells as well. We carried out the miR-198 mimic and circPTCD3 overexpressing vector transfection. The miR-198 mimic transfection apparently augmented the level of miR-198; on the other hand, the co-transfection with miR-198 mimic and circPTCD3 overexpressing vector lowered the level of miR-198 in comparison with that in the miR-198 group (Figure 4A). By means of the function studies, we, at first, discovered that miR-198 attenuated not only the cell proliferation but also the migration and colony formation of breast cancer cells. Together with that, circPTCD3 reversed these functions of miR-198 partially, further indicating the relationship existing between circPTCD3 and miR-198 (Figure 4B–D).
CircPTCD3 Promoted the Growth of Breast Cancer Cells in vivo
Thereafter, we carried out the in vivo study to further investigate the biological function of circPTCD3 in the progression of breast cancer. Stable MCF-7 cell line was established with the transfection of sh-circPTCD3 or circPTCD3 vectors and their control, correspondingly. Both the size and weight of the tumours in the circPTCD3 were observed as higher in comparison with the control tumours. Contrarily, the circPTCD3 deletion lowered the tumour size and weight of tumours in comparison with the control group (Figure 5A–C). IHC assay, detecting the expression of ki67, the proliferation biomarker, was also carried out. In accordance with the expectations, circPTCD3 overexpression promoted the expression of ki67 while the knock-down of circPTCD3 inhibited that of ki67 (Figure 5D).
Discussion
CircRNAs have been fully documented as playing critical roles in the progression of breast cancer such as circRNF20, circular RNA Foxo3, circKDM4C, hsa_circ_001783, circ-DNMT1 and CircIRAK3.11–16 In the current study, we figured out an novel circRNA, termed as circPTCD3. We illustrated the fact that circPTCD3 originated from exons 3 to 9 of PTCD3, together with forming a loop structure by means of connecting the 3ʹ and 5ʹ splice sites. The stability of circPTCD3 was confirmed by its stable expression subjected to the RNase R digestion. To the best of our understanding, this is the first study dealing with the circPTCD3 emphasizing its modified expression as well as biological impact. The up-regulated expression and stability of circPTCD3 make it a potential biomarker as a diagnostic and therapeutic target.
Previous studies have demonstrated diverse mechanisms underlying the effect of circRNAs, such as acting as scaffolds of protein complexes, regulating protein subcellular localization, sequestering RNA-binding proteins, or functioning as the sponge of miRNAs. In the present study, we focus on the ability of circPTCD3 sponging miRNAs.
Plenty of miRs have been demonstrated as participating in the biological process of breast cancer. MiR-137, miR-17, miR-186-3p, miR-454-3p and miR-29a inhibited not only the proliferation but also the migration and invasion of human breast cancer cells by means of targeting the DUSP4, JAB1, EREG, Wnt/β and SUV420H2, correspondingly.17–20 Among them, miR-198, belonging to the miR-198 family, have been extensively investigated for playing a crucial role in the cellular differentiation and development. As in cancers, a number of research works shed light on the oncogenetic or anticancer function of miR-198, which include ovarian cancer, myeloid leukaemia, colorectal cancer, non-small cell lung cancer and gastric cancer. However, the role of miR-198 in the progression of breast cancer is limited. miR-198 has been demonstrated to targetLINC00473 and MAPK1 in breast cancer cells.21 In addition to that, miR-198 was reported to act as an anticancer gene in breast cancer by targeting CUB domain-containing protein 1.22 In the current research work, miR-198 was determined to be the target miRNA of circPTCD3, which was down-regulated in the breast cancer tissues and cells. Together with that, the function study revealed the anticancer effect of miR-198 in breast cancer, which is consistent with the previous study. Interestingly, miR-198 overexpression reversed the impact of circPTCD3, further verifying the relationship existing between them.
MiRNAs target mRNAs to induce the translation inhibition or degradation of them.23 The limitation of this study is that we have not figure out the downstream target gene of miR-198 in breast cancers. It will be carried out in the future.
To conclude, our findings shed light on the fact that circPTCD3 promotes the malignant behaviour of breast cancer by means of sponging miR-198, which indicates its potentiality to be a diagnostic or therapeutic target of breast cancer.
Disclosure
All authors declare that no conflicts of interest exist in the research.
References
1. Woolston C. Breast cancer. Nature. 2015;527(7578):S101. doi:10.1038/527S101a
2. Cook DJ, Kallus J, Jornsten R, Nielsen J. Molecular natural history of breast cancer: leveraging transcriptomics to predict breast cancer progression and aggressiveness. Cancer Med. 2020;9(10):3551–3562. doi:10.1002/cam4.2996
3. Khandekar D, Tiriveedhi V. Role of BET inhibitors in triple negative breast cancers. Cancers. 2020;12(4):784. doi:10.3390/cancers12040784
4. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer Tokyo. 2018;25(1):1–7. doi:10.1007/s12282-017-0793-9
5. Agostini M, Ganini C, Candi E, Melino G. The role of noncoding RNAs in epithelial cancer. Cell Death Discov. 2020;6(1):13. doi:10.1038/s41420-020-0247-6
6. Wei L, Sun J, Zhang N, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19(1):62. doi:10.1186/s12943-020-01185-7
7. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
8. Hua JT, Chen S, He HH. Landscape of noncoding RNA in prostate cancer. Trends Genet. 2019;35(11):840–851. doi:10.1016/j.tig.2019.08.004
9. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. doi:10.1055/s-0034-1397344
10. Lou W, Ding B, Fu P. Pseudogene-derived lncRNAs and their miRNA sponging mechanism in human cancer. Front Cell Dev Biol. 2020;8:85. doi:10.3389/fcell.2020.00085
11. Wu J, Jiang Z, Chen C, et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–192. doi:10.1016/j.canlet.2018.05.033
12. Du WW, Yang W, Li X, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37(44):5829–5842. doi:10.1038/s41388-018-0369-y
13. Liu Z, Zhou Y, Liang G, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10(2):55. doi:10.1038/s41419-018-1287-1
14. Liang Y, Song X, Li Y, et al. circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR-548p/PBLD axis in breast cancer. Oncogene. 2019;38(42):6850–6866. doi:10.1038/s41388-019-0926-z
15. Cao L, Wang M, Dong Y, et al. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1alpha/HK2. Cell Death Dis. 2020;11(2):145. doi:10.1038/s41419-020-2336-0
16. Huang W, Yang Y, Wu J, et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-beta signalling. Cell Death Differ. 2019.
17. Min J, Han TS, Sohn Y, et al. microRNA-30a arbitrates intestinal-type early gastric carcinogenesis by directly targeting ITGA2. Gastric Cancer. 2020;23(4):600–613. doi:10.1007/s10120-020-01052-w
18. He M, Jin Q, Chen C, et al. The miR-186-3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in breast cancer cells. Oncogene. 2019;38(28):5551–5565. doi:10.1038/s41388-019-0817-3
19. Ren L, Chen H, Song J, et al. MiR-454-3p-Mediated Wnt/beta-catenin Signaling Antagonists Suppression Promotes Breast Cancer Metastasis. Theranostics. 2019;9(2):449–465. doi:10.7150/thno.29055
20. Wu Y, Shi W, Tang T, et al. miR-29a contributes to breast cancer cells epithelial–mesenchymal transition, migration, and invasion via down-regulating histone H4K20 trimethylation through directly targeting SUV420H2. Cell Death Dis. 2019;10(3):176. doi:10.1038/s41419-019-1437-0
21. Niu L, Zhou Y, Zhang W, Ren Y. Long noncoding RNA LINC00473 functions as a competing endogenous RNA to regulate MAPK1 expression by sponging miR-198 in breast cancer. Pathol Res Pract. 2019;215(8):152470. doi:10.1016/j.prp.2019.152470
22. Hu Y, Tang Z, Jiang B, Chen J, Fu Z. miR-198 functions as a tumor suppressor in breast cancer by targeting CUB domain-containing protein 1. Oncol Lett. 2017;13(3):1753–1760. doi:10.3892/ol.2017.5673
23. Caiment F, Gaj S, Claessen S, Kleinjans J. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Res. 2015;43(5):2525–2534. doi:10.1093/nar/gkv115
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.