Back to Journals » Drug Design, Development and Therapy » Volume 15
A New Strategy for the Rapid Identification and Validation of the Direct Targets of Aconitine-Induced Cardiotoxicity
Authors Wei J, Fan S , Yu H, Shu L, Li Y
Received 22 August 2021
Accepted for publication 28 October 2021
Published 13 November 2021 Volume 2021:15 Pages 4649—4664
DOI https://doi.org/10.2147/DDDT.S335461
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Jinxia Wei,* Simiao Fan,* Hongxin Yu,* Lexin Shu, Yubo Li
School of Chinese Materia Medica, Tianjin University of Traditional Chinese Medicine, Tianjin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yubo Li
School of Chinese Materia Medica, Tianjin University of Traditional Chinese Medicine, No. 10, Poyang Lake Road, West Zone, Tuanbo New City, Jinghai District, Tianjin, 301617, People’s Republic of China
Tel +86-22-59591974
Email [email protected]
Background: The interaction of small molecules with direct targets constitutes the molecular initiation events of drug efficacy and toxicity. Aconitine, an active compound of the Aconitum species, has various pharmacological effects but is strongly toxic to the heart. The direct targets of aconitine-induced cardiotoxicity remain unclear.
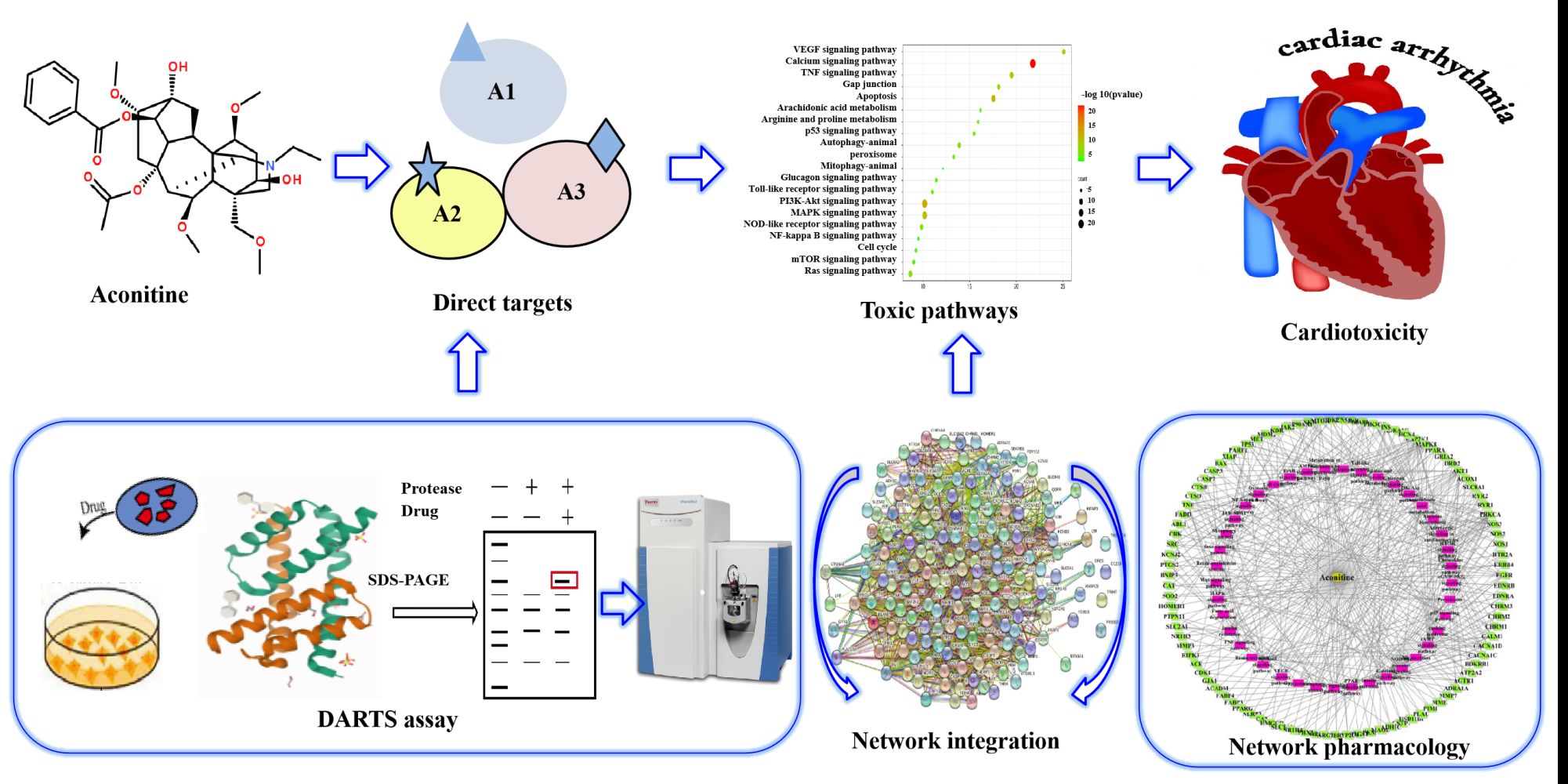
Methods: We predicted the toxic targets of aconitine based on network pharmacology and followed a novel proteomic approach based on the “drug affinity responsive target stability” technology combined with LC-MS/MS to identify the direct targets of aconitine. The identified targets were analysed from the perspective of multilevel and multidimensional bioinformatics through a network integration method. The binding sites were investigated via molecular docking to explore the toxicity mechanism and predict the direct targets of aconitine. Finally, atomic force microscopy (AFM) imaging was performed to verify the affinity of aconitine to the direct targets.
Results: PTGS2, predicted by network pharmacology as a toxic target, encodes cyclooxygenase 2 (COX-2), which is closely related to myocardial injury. Furthermore, cytosolic phospholipase A2 (cPLA2) is the upstream signal protein of PTGS2, and it is a key enzyme in the metabolism of arachidonic acid during an inflammatory response. We determined cPLA2 as a direct target, and AFM imaging verified that aconitine could bind to cPLA2 well; thus, aconitine may cause the expression of PTGS2/COX-2 and release inflammatory factors, thereby promoting myocardial injury and dysfunction.
Conclusion: We developed a complete set of methods to predict and verify the direct targets of aconitine, and cPLA2 was identified as one. Overall, the novel strategy provides new insights into the discovery of direct targets and the molecular mechanism of toxic components that are found in traditional Chinese medicine.
Keywords: aconitine, cardiotoxicity, direct targets, drug affinity responsive target stability technology, atomic force microscopy
Graphical Abstract:
Introduction
Aconitum is a type of traditional Chinese medicine (TCM) in the Ranunculaceae family with a long medicinal history in China. Aconitums, represented by Aconiti Lateralis Radix Praeparata, Aconite Radix, and Aconiti Kusnezoffii Folium, have high medicinal value.1 Specifically, Aconitums cure rheumatism and show anti-inflammatory and analgesic properties; thus, they are highly regarded by physicians and pharmacists of various dynasties.2 Modern pharmacological investigations have revealed that Aconitum species possess various therapeutic effects, such as anti-inflammatory, analgesic, antitumour, cardiotonic, and vasodilative effects.3 Aconitine is an alkaloid, representative of those in Aconitums, that has significant therapeutic effects and toxicity simultaneously. Specifically, it shows various pharmacological effects, such as analgesic, cardiotonic, antitumour, and immunosuppressive effects.4 However, countless poisoning cases caused by aconitine have been reported in recent years.5 If it is used irrationally and the therapeutic window is exceeded, aconitine can cause cardiotoxicity. Furthermore, aconitine can cause vagus nerve stimulation, promote the release of acetylcholine, reduce the self-regulation of the sinoatrial node, and cause sinus bradycardia. In severe cases, it can cause sinus arrest and seriously damage the heart system, which greatly limits the clinical application of Aconitum species.6 The mechanisms of aconitine-induced cardiotoxicity are multifaceted, and aconitine can cause different degrees of heart damage by affecting ion channels, damaging DNA, affecting energy metabolism, and causing cell apoptosis.7–10 However, the direct target of aconitine is still unclear and research on the initial events of toxicity is not thorough enough. Hence, identifying the key targets of aconitine and revealing the underlying mechanism of aconitine-induced cardiotoxicity has become a critical issue in the toxicity research and clinical application of aconitine.
The target protein is an irreplaceable starting point for drugs to exert therapeutic or toxic effects in vivo. Small compounds reach the target organs and interact with direct target proteins, which signifies the molecular initiation event of their efficacy or toxicity. The key and difficult point in this study was the discovery of the direct target proteins of small molecules. Determining the direct targets of active compounds is needed to explain the pharmacological action and toxic effects of herbal medicine; however, this is a difficulty faced in the current research on Chinese medicine.11 At present, there are two main strategies for the discovery and identification of drug direct targets, viz. virtual screening and experimental research. Virtual screening has the advantages of being intuitive, convenient, and efficient, which can significantly shorten the experimental cycle, reduce research costs, and improve the success rate of target screening. Virtual screening based on network pharmacology has high throughput and is easy to operate. Moreover, this method is sensitive and does not need to consider the content of small molecules, which could be used for the discovery of most drug targets.12 TCM network pharmacology integrates system biology and other disciplines, and it explores the relationship between drugs and complex diseases from a holistic perspective; it has the advantages of integrity, synergy, and dynamics, which complement the basic characteristics of TCMs.13 Furthermore, it is important in the screening of potential effective or toxic compounds as well as the research of target proteins.14
With the development and progress of chemical biology, experimental research on the direct targets of small molecules currently includes click chemistry (CC), drug affinity responsive target stability (DARTS), cellular thermal shift assay (CETSA) and so on. The CC strategy entails the design of small-molecule probes to reflect changes in the functional state of target proteins in cells. Through this method, it is necessary to introduce an alkynyl group to small molecules and then fish the corresponding targets after modification.15 However, the CC strategy is not suitable for molecules which are not easily modified or whose activity changes after binding with an alkynyl group. The principle of the DARTS assay is that the protein structure changes in the direction of energy reduction and becomes more stable after binding with small molecules to resist protease digestion. The DARTS technology does not require any chemical modification of compounds and target proteins; it is easy to operate and can be used together with proteomic analysis to identify unknown target proteins of most small molecules.16,17 Furthermore, the binding of small molecules to the targets must be verified after identifying the direct targets. Some verification methods, such as molecular docking, surface plasmon resonance, atomic force microscopy (AFM), and siRNA interference, can be used to explore the optimal binding mode of small molecules and targets.18,19
In this study, we first predicted the potential toxic targets of aconitine based on network pharmacology and subsequently identified the direct targets using a novel proteomic approach based on DARTS and LC-MS/MS. The potential direct targets and toxicity mechanisms of aconitine were explored through network integration analysis. The binding sites between aconitine and its direct targets were investigated via molecular docking analysis. Finally, the affinity of aconitine to the target protein was verified via an AFM assay, thereby forming a complete set of direct target exploration and verification systems (Figure S1). The new strategy predicted several direct targets of aconitine and explained the potential mechanism of aconitine-induced cardiotoxicity. This study not only provided a theoretical foundation for the safe and effective clinical use of aconitine but also inspired a new perspective for research on the toxicity mechanism of TCMs.
Materials and Methods
Materials and Reagents
Aconitine (302-27-2, purity > 98%) was purchased from Chengdu Herbpurify Co., Ltd. (Chengdu, China). Foetal bovine serum (FBS), Dulbecco’s Modified Eagle Medium (DMEM), trypsin, and a penicillin–streptomycin mixed solution (100× double antibody) were obtained from Gibco (Gaithersburg, MD, USA). Dimethyl sulfoxide (DMSO) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO, USA). The recombinant human protein cytosolic phospholipase A2 (cPLA2) was obtained from Shanghai Kanglang Biotechnology Co., Ltd. (Shanghai, China). Phosphate-buffered saline (PBS) (1×, pH=7.2–7.4), M-PER cell lysate, and other reagents were obtained from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China). A pure water system (Thermo Fisher Scientific, USA) and a 5415D small high-speed centrifuge (Eppendorf, Germany) were used. The LC-MS/MS analysis was performed using a quadrupole Orbitrap mass spectrometer coupled with liquid chromatography (Thermo Scientific, San Jose, CA, USA). The data files were searched using MASCOT software (Matrix Science, Boston, MA, USA) to obtain qualitative identification information of the target proteins and peptide molecules. Discovery Studio software (2017 R2 client) was used for molecular docking to explore the interaction between aconitine and its direct target proteins. Finally, a Bruker Dimension® IconTM atomic force microscope (Bruker, Germany) was used.
Network Pharmacology Analysis
Target Prediction of Aconitine
The 3D structure of aconitine was acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Target genes of aconitine were obtained by importing the 3D structure of aconitine into the SwissTargetPrediction (http://www.swisstargetprediction.ch/) and PharmMapper (http://lilabecust.cn/pharmmapper/) databases. The gene official symbol format was acquired using the UniProt database (https://www.uniprot.org/). Finally, the predicted target genes of aconitine were obtained by merging and deleting duplicate items.
Acquisition of Target Genes Related to Aconitine-Induced Cardiotoxicity
Information about cardiotoxicity-associated target genes was searched in the CTD (http://ctdbase.org), TTD (http://bidd.nus.edu.sg/group/cjttd/), Drugbank (https://www.drugbank.ca/), Genecards (https://www.genecards.org/), and OMIM (http://www.omim.org/) databases after entering the keywords “Myocardial injury”, “Cardiotoxicity”, and “Arrhythmia”. The obtained targets were normalised using the Retrieve/ID mapping of the UniProt database to obtain their official symbols. Next, the potential target genes were identified using the Venn online platform. In addition, the reported target genes were collected from PubMed, CNKI, Web of Science, and other databases. After merging and deduplication, the potential core target genes were identified.
Construction of Protein–Protein Interaction Network Between Core Targets
The main function of the STRING database (https://string-db.org/) is to provide physical and functional protein-protein association data and construct the relationship between targets. The protein-protein interaction (PPI) network was constructed by inputting the potential core target genes in the STRING database. Then, the TSV format of the PPI network was saved and imported into the Network Analyser module of Cytoscape 3.6.1 to construct and analyse the target network relationship. The correlation degree among common targets of drug–disease was analysed based on the degree of nodes, and the key targets in the PPI network were screened via topological analysis and number sequencing. Finally, a PPI network diagram was constructed by defining the node size and degree centrality of colour-sort reflection.
Gene Ontology and KEGG Pathway Enrichment Analysis
To investigate the biological function of aconitine on potential target genes related to cardiotoxicity, we imported the candidate targets into the Metascape database for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. GO terms and KEGG pathways with p < 0.05 were considered statistically significant. GO analysis divides gene function into three parts: cellular component, molecular function, and biological process, which screens out pathways related to targets. The pathway database in the KEGG database integrates the functional information of genes, proteins, and metabolites. The top 20 relevant KEGG pathways were visualised as bubble plots using the Omicshare cloud platform. To analyse the association among aconitine, candidate targets, and cardiotoxicity-related pathways, an “aconitine–targets–pathways” network was constructed using Cytoscape 3.6.1. The network diagram plotted in Figure 1E visually displays the multidimensional regulation mechanism of aconitine-induced cardiotoxicity in a complex system.
Study on H9c2 Myocardial Cell Injury Induced by Aconitine
H9c2 Cardiomyocyte Culture and Drug Treatment
The H9c2 cardiomyocyte cell line was purchased from Beijing Beina Chuanglian Biotechnology Research Institute (Beijing, China), and the relevant experiments were approved by the Ethics Committee of Tianjin University of Traditional Chinese Medicine. The cells were cultured in DMEM containing 100 U/mL penicillin and 100 μg/mL streptomycin supplemented with 10% FBS. The cells were kept in a cell incubator at 37 °C with 5% CO2. Aconitine was dissolved in DMSO to prepare a stock solution with a concentration of 10 mM, which was stored in a refrigerator at −20 °C after subpackaging and was diluted with DMEM before use. H9c2 cells in the logarithmic growth phase were digested with 0.25% trypsin, resuspended in DMEM, seeded on 96-well plates (approximately 3×104 cells), and cultured in a 5% CO2 incubator at 37 °C for 24 h. The cell groups were divided into solvent control, normal, and aconitine treatment groups. After incubation for 24 h, the culture medium was removed. The solvent control group was then replaced with a culture medium containing 3% DMSO, the normal group was replaced with a normal culture medium, and the aconitine-treatment group was replaced with 100 µL of a culture medium with aconitine concentrations of 50, 100, 150, 200, 250, 300, and 400 μmol/L. Each group was incubated for 24 h.
Cell Viability Detection via MTT Assay
The cell viability of each group was determined based on the MTT assay. After discarding the medium, 10 μL of MTT reagent (5 mg/mL) and 90 μL of DMEM were added into each well of 96-well plates, and the cells were further cultured at 37 °C for 4 h. Then, 150 μL DMSO was added to each well to dissolve the crystallisation after removing the medium. The absorbance values were read at 570 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
DARTS Assay
Extraction and Quantification of H9c2 Cardiomyocyte Protein
After discarding the medium, the cells were washed with sterile PBS 1–2 times, and 0.25% EDTA-trypsin solution was added to digest the cells. PBS was added to resuspend the cells after centrifugation at 100 rpm for 5 min; subsequently, the supernatant was discarded. Then, M-PER cell lysate (containing phosphatase and protease inhibitor) was added and placed on ice for 30 min. The cell lysate was centrifuged at 12,000 rpm for 15 min at 4 °C, and the supernatant was transferred into clean tubes. The protein concentration was measured in accordance with the manufacturer’s instructions using a BCA protein concentration assay kit (Solarbio, Beijing, China).
Selection of Types and Concentrations of Protease
A key point of the DARTS technique is the addition of an appropriate protease. Subtilisin, a thermophilic protease, and Streptomyces protease can be applied to DARTS technology, with Streptomyces protease being recommended by the inventor of the DARTS technology.20–23
The samples were divided into normal and aconitine groups. Neither aconitine nor enzymes were added to the normal group. For the aconitine group, three concentration levels (100 μM, 200 μM, and 400 μM) were set, which covered the toxicity concentration of the cell viability test. Fifty microliters of protein solution were added to each group. After incubating for 1 h, 2 μL of the corresponding proportion of the digestive enzyme mixture pronase (1:100, 1:500, 1:1000, 1:3000) was added to the aconitine group and mixed. Two microliters of 1× TNC buffer were added to the normal group. After digestion for 30 min at 25 °C, 0.5 M EDTA (pH 8.0) protease inhibitor was added to terminate the enzyme reaction. Finally, loading buffer was added and mixed using a vortex. The protein sample was boiled for 5 min in a 95 °C water bath to denature the protein. The effect of the protease was verified via sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE).
Co-Incubation of Aconitine with Proteins
The H9c2 protein solution in the aconitine group was supplemented with varying concentrations of aconitine (100 μM, 200 μM, and 400 μM) and allowed to incubate for 1 h at room temperature. In addition, the negative control group was set, and the protein solution was incubated with the same concentration of DMSO. Then, 2 μL of pronase solution (1:100) was added to all experimental groups except the normal group for 30 min digestion at 25 °C. Finally, the reaction was terminated on ice, loading buffer was added to the samples, and they were boiled and analysed as mentioned above.
Electrophoresis and Identification of Differential Proteins
The total protein sample was separated on a 12% gel via SDS-PAGE and stained with a Coomassie Brilliant Blue staining solution, and then decolourized until the blue background was removed. The differential protein bands in the aconitine group were analysed using the Odyssey gel imaging system (LI-COR Biosciences, USA). After reduction and alkylation, trypsin was used to hydrolyse the differentially expressed proteins. Finally, LC-MS/MS was used to analyse the samples. The qualitative information of the target proteins was obtained using the MASCOT software.
Integration Analysis of Potential Targets of Aconitine
In this study, we performed a network integration analysis of the targets predicted by network pharmacology and the DARTS technology. The potential targets were imported into the STRING database. The PPI network of the target genes was generated from the STRING database, which helps mine the relationship between targets. The direct targets of aconitine were discovered via network analysis as well as literature research.
Molecular Docking
The 3D structure of aconitine was searched and downloaded from the PubChem database. Using the PDB database (http://www.rcsb.org/), we acquired the three-dimensional crystal structures of proteins (cPLA2 [PDB: 1CJY], ORC5 [PDB: 5UI7], CTNNA1 [PDB: 4EHP], and GYS1 [PDB: 3CX4]) containing ligands with high resolution. The proteins and aconitine were prepared using the Discovery Studio software (2017 R2 client) for the molecular docking study. Before the molecular docking operation, it is necessary to verify whether the selected docking procedure and parameters are suitable for the structure of the receptor protein. The semi-flexible docking mode was selected for the molecular docking analysis.
AFM Analysis
Recombinant human protein cPLA2 (50 μg) was dissolved and diluted with 1× PBS (pH 7.2–7.4) to obtain a 400 μmol/L solution. A PBS solution containing 0.1% DMSO was used to prepare the aconitine solution at a concentration of 400 μmol/L. The protein solution was then mixed with the aconitine solution at a ratio of 1:1 and incubated for 30 min in a water bath at 37 °C. Ten microliters of the resulting mixture and 10 μL of protein solution were dropped onto a glass slide and placed at room temperature to prepare the crystalline sample. Then, the crystals of the protein and protein-aconitine complex were scanned via ICON AFM in tapping mode. The morphology, distribution, aggregation degree, and other morphological characteristics of protein crystals on each slide were recorded. The crystal images were processed using NanoScope Analysis 1.8.
Statistical Analysis
All experimental data are presented as mean ± SD. Statistical analyses were conducted using the SPSS software (version 13.0). One-way analysis of variance followed by Tukey’s post hoc test was used for two-group comparisons, and statistical significance was set at p < 0.05.
Results
Acquisition of Common Target Genes Related to Aconitine and Cardiotoxicity and PPI Network Analysis
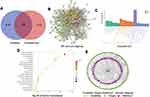
By searching the Swiss Target Prediction and PharmMapper databases, 526 target genes of aconitine were acquired based on structural similarity and pharmacophore modelling. Similarly, 1241 cardiotoxicity-related target genes were collected from the CTD, TTD, Drugbank, Genecards, and OMIM databases. The overlapping targets were obtained using the Venn online platform; 108 overlapping potential target genes were identified. The Venn diagram is shown in Figure 1A. In addition, 46 reported target genes (Table S1) were identified by searching databases. A total of 152 core targets were obtained after removing the repetitive targets. Furthermore, a PPI network diagram (Figure 1B) was constructed using the STRING database to clarify the relationship between core targets. Meanwhile, using the Cytoscape 3.6.1 software, we screened out the top 14 target genes (ALB, AKT1, TNF, TP53, CASP3, EGFR, MAPK8, MAPK1, SRC, CAT, PTGS2, CYCS, HSP90AA1, MTOR) with the highest degree value by analysing the topological parameters and correlation between targets. Among them, PTGS2, also known as cyclooxygenase 2 (COX-2), is upregulated when myocardial cells are stimulated by inflammation. The upregulation of COX-2 and the synthesis of prostaglandins promote the release of MMPs and then regulate collagen metabolism and the degree of fibrosis in myocardial tissue, which is related to myocardial diseases.24
GO and KEGG Pathway Enrichment Analysis
To further reveal the related pathways and mechanisms of aconitine-induced cardiotoxicity, the KEGG pathway and GO enrichment analysis of 152 core targets were conducted using the Metascape database. Specifically, the GO functional enrichment analysis showed that aconitine-induced cardiotoxicity is mainly involved in circulatory and muscle system processes, as well as in the regulation of blood circulation, transmembrane transport, and ion transport. The enrichment results were ranked based on the p-value, and the integrated GO analysis results are shown in Figure 1C. KEGG enrichment analysis showed that 152 targets were mainly concentrated in 181 pathways (p < 0.05), of which 41 were closely related to cardiotoxicity, such as the VEGF, calcium, TNF, MAPK, PI3K-Akt, and mTOR signalling pathways, as well as gap junction, apoptosis, and metabolism of arachidonic acid. The top 20 pathways are shown in Figure 1D. Meanwhile, the network diagram of “aconitine–targets–pathways” (Figure 1E) was obtained using the Cytoscape 3.6.1 software, in which the nodes are represented by different colours and shapes; yellow represents aconitine, green represents the targets, purple represents the signalling pathways, and the edges represent the relationships between nodes. The network contained 137 nodes, including aconitine, 95 targets related to cardiotoxicity, and 41 pathways, which intuitively reveal the characteristics of multiple targets and multiple pathways of aconitine.
Toxicity of Aconitine on H9c2 Cardiomyocytes
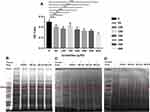
Aconitine has a definite effect on mitochondrial dehydrogenase.25 Therefore, the MTT assay was applied to detect the effect of aconitine on cell viability and screen the toxic concentration to lay a foundation for fishing direct targets of aconitine. The viability of cells treated with different concentrations of aconitine is shown in Figure 2A. Compared with that in the normal group, the cell viability in the aconitine-treated group significantly decreased as the concentration increased (p < 0.01); this indicated that aconitine inhibited the viability of H9c2 cardiomyocytes in a concentration-dependent manner. The MTT assay revealed that the inhibition rate of aconitine on H9c2 cardiomyocytes was approximately 50% at a concentration of 400 μM. Hence, 400 μM was selected as the experimental concentration for the follow-up study.
Direct Target Identification Based on DARTS Technology
The extraction of protein from cells is a key step in fishing the direct targets of aconitine in the DARTS technology. However, the lysates used in the process of protein extraction directly affect the sensitivity and resolution of subsequent experiments.26 In this study, protein was extracted from three bottles (75 cm2) of cells by adding 500 μL M-PER lysate. A 5 μg/μL protein solution was obtained, which met the requirements of the DARTS experiment. In addition, the selection of a protease is a key operation in the DARTS experiment. Pronase was selected for this experiment, and the optimal digestion time was determined to be 30 min. The results of the investigation of enzyme concentrations (pronase-to-protein ratios: 1:100, 1:500, 1:1000, and 1:3000) are shown in Figure S2. The digestion ability of pronase-to-protein ratios 1:100 and 1:500 was higher than that of the 1:1000 and 1:3000 ratios. The protein bands were clear in the normal group and became clearer when the enzyme concentration decreased. Thus, a 1:100 pronase-to-protein ratio was used for subsequent experiments. In this case, the interference bands were smaller, which could help detect differential protein bands. Then, DARTS was performed in accordance with reported protocols.27 After incubation with aconitine for 1 h, the protein was digested with 1:100 pronase to protein for 30 min. The whole protein bands obtained via SDS-PAGE and Coomassie Brilliant Blue staining are shown in Figure 2B–D. Furthermore, the differential protein bands were compared to the negative control group. The protein band of the aconitine group was significantly deeper than that of the negative control group at 70 kDa, indicating that the protein band had a specific affinity with aconitine and thus may be the target protein of aconitine. Therefore, the differential band was accurately cut for further enzymolysis and LC-MS/MS analysis (Figure S3). By comparing the protein mass spectrometry data between the normal and negative control groups, proteins with molecular masses of approximately 70 kDa were screened. The peptide sequence was identified from the protein database, and 15 proteins of approximately 70 kDa were identified (Table 1). The content of 15 target proteins in the negative control group decreased sharply after the addition of protease. Aconitine increased the content of the proteins in a dose-dependent manner, which indicated that it can specifically bind proteins and improve their stability.
 |
Table 1 The Information of Direct Target Proteins Fished by DARTS Technology |
Integration Analysis of Direct Targets of Aconitine
In this study, 152 core targets predicted by network pharmacology and 15 direct targets from the DARTS experiment were imported into the STRING database for protein interaction analysis (Figure 3). According to literature reviews and enrichment analysis of the KEGG function, the direct targets cPLA2, GYS1, CTNNA1, and ORC5 interact with network pharmacological targets and are related to myocardial injury.
 |
Figure 3 PPI network diagram of 152 core targets from network pharmacology and 15 direct targets from DARTS. |
Glycogen synthase (GYS) is the rate-limiting enzyme in glycogen synthesis, and it includes glycogen synthase 1 (GYS1) and 2 (GYS2). Shi et al found that GYS1-mediated glycogen accumulation promotes FLS-mediated synovial inflammation in rheumatoid arthritis by blocking the activation of AMPK, thus demonstrating that glycogen metabolism is associated with chronic inflammation.28 Glycogen is the storage of glucose in many mammalian tissues. The ability to synthesise glycogen in cardiac muscles is crucial for the healthy development of the heart. Impaired cardiac glycogen synthesis could be a significant contributor to congenital heart disease.29 In addition, Zhang et al found that aconitine can inhibit the energy metabolism of cardiomyocytes by reducing the contents of glycogen, succinate dehydrogenase, and cytochrome c oxidase in cells.30 Recently, CTNNB1 gene levels were found to play an important rate-limiting role in the regulation of insulin secretion. Thus, researchers have investigated whether the binding partner CTNNA1 also plays a role in this process. The results showed that CTNNA1-mediated actin remodelling may be involved in the regulation of insulin secretion.31 The origin recognition complex (ORC) is encoded by the latheo gene, it consists of six isoforms, (ORC1–ORC6), and it is involved in DNA biosynthesis in the cell cycle pathway. ORC5 has been associated with hepatocellular carcinoma and colon cancer; thus, it is a new candidate biomarker for the survival monitoring of patients with hepatocellular carcinoma.32 Furthermore, cPLA2 has a wide range of biological activities, such as playing a leading role in arachidonic acid release, initiating prostaglandin signalling cascade, participating in membrane phospholipid remodelling, and regulating inflammation and various related physiological and pathological processes.33 In the failing heart, sPLA2 migrates into the myofibrillar membrane, whereas cPLA2 accumulates in the cytoplasm.34
Molecular Docking
Based on the target proteins that overlapped between PPI and the KEGG network topology analysis, we tested the docking precision between aconitine and the following potential direct targets: cPLA2 (PDB: 1CJY), GYS1 (PDB: 3CX4), ORC5 (PDB: 5UI7), and CTNNA1 (PDB: 4EHP). Method validation must be performed before the molecular docking of aconitine and proteins. The root-mean-square deviation (RMSD) value was used as the evaluation standard. For RMSD < 2.0 Å, the smaller the value, the more accurate the docking result. The RMSD values of the four direct target proteins, cPLA2, GYS1, ORC5, and CTNNA1, were 1.1874 Å, 0.7458 Å, 1.3682 Å, and 0.4265 Å, respectively, which are less than 2.0 Å. Thus, the method selected in this study is reliable and can be used in follow-up virtual screening research. Molecular docking was carried out using the CDocker module in the Discovery Studio software. CDocker is a semi-flexible docking program based on the CHARMM force field. Virtual computer screening was performed using this program. After docking, we checked the docking scores (cdocker-interaction-energy) and predicted the affinity and binding mode of drugs and proteins. A higher score suggested stronger binding activity of the ligand with the receptor. Based on the CDocker interaction energy scores (Table S2), aconitine had a good binding activity with four potential direct proteins (cPLA2, GYS1, CTNNA1, and ORC5); 3D and 2D diagrams present the molecular docking models of aconitine with those direct target proteins (Figure 4). The receptor–ligand interactions primarily included van der Waals forces and hydrogen bonds. For example, aconitine could match with cPLA2 by forming a stable hydrogen bond at the HIS62 and ASN64 sites (Figure 4A). Meanwhile, aconitine produced various degrees of van der Waals forces with ASN95, ALA94, and ASN65 amino acid residues in cPLA2 and formed hydrocarbon bonds with TYR96 and ASN64. The results indicated that aconitine is in a reasonable cavity of cPLA2, and it can stably bind to this receptor protein. Similar to the case of cPLA2, the combination of aconitine with GYS1 revealed both van der Waals forces and hydrogen bond interactions. However, the hydrogen bonding sites were ARG300, HIS96, and ASP331, whereas the sites of the van der Waals interactions were HIS161, ASP137, and THR16 (Figure 4B). In addition, the benzene ring of aconitine formed pi–cation and pi–alkyl interactions with HIS139 and TYR165 residues, respectively. Moreover, CTNNA1 bound to aconitine mainly by forming hydrogen bonds with SER252, THR251, and GLU128. Aconitine also interacted with the receptor through van der Waals forces produced by LEU124 and other amino acid residues (Figure 4C). ORC5 combined with aconitine through two stable hydrogen bonds with GLU502 and ALA542 and various van der Waals forces with amino residues, such as VAL535 and PRO505. In addition, alkyl interactions existed between the ligand and residues ILE689 and LEU723 (Figure 4D). These data suggest that the four target proteins can be considered direct targets of aconitine-induced cardiotoxicity.
cPLA2 as a Potential Direct Target of Aconitine
To further screen the direct targets of aconitine-induced cardiotoxicity, literature investigations were carried out from the perspective of biological pathways. The direct target cPLA2 was found to be the upstream signal protein of key target PTGS2 predicted by network pharmacology; it is also an important starting material and key metabolic enzyme of arachidonic acid metabolism. The molecular docking results showed that aconitine has a strong binding ability to the active cavity of cPLA2. The metabolism of arachidonic acid by cyclooxygenase (COX-1 and COX-2) is a key step in the synthesis of prostaglandins, such as PGI2, PGD2, and PGE2, which are also the most reported metabolic pathways of arachidonic acid. Arachidonic acid and its metabolite-mediated inflammation are associated with the occurrence and development of myocardial injury, and they can be enriched in the KEGG pathway of network pharmacology. Furthermore, cPLA2 and sPLA2 isozymes were detected in the cytoplasm of normal cardiomyocytes by Mchowat et al. In the failing heart, sPLA2 migrated into the myofibrillar membrane, whereas cPLA2 accumulated in the cytoplasm.34 PTGS2 gene encodes COX-2, whose expression increases when cells are stimulated by inflammation; this is a decisive factor of inflammation-mediated cytotoxicity.35 Bolli et al found that COX-2 and its metabolites increased following 24 h ischemia in a rabbit ischemia/reperfusion injury model.36 Saito et al found that strong immune activity of COX-2 can be detected in myocardial cells after acute myocardial infarction. The addition of a COX-2 inhibitor can significantly reduce the infarct size and improve myocardial contractility. These results showed that the expression of COX-2 promotes myocardial injury and dysfunction.37 COX-2 is an important rate-limiting enzyme that catalyses the conversion of arachidonic acid to PGE2. PGE2 is an effective mediator of inflammation, and it is widely involved in ischemia-induced neuronal damage, inflammatory response, and neurodegenerative diseases.38 Aconitine has a two-way regulatory effect on anti-immunosuppression. At low dosages, it promotes the expression of PEG2 via the PEG2/COX-2 signalling pathway.39 Based on comprehensive analysis, we speculate that aconitine induces cardiotoxicity by activating the direct target cPLA2, mediating the release of arachidonic acid, and promoting the release of inflammatory factors via the PGE2/COX-2 pathway. Therefore, cPLA2 may be a potential direct target of aconitine-induced cardiotoxicity. The potential mechanism of aconitine toxicity is shown in Figure 5.
 |
Figure 5 Potential toxicity mechanism of aconitine by acting on direct target cPLA2. |
AFM Analysis
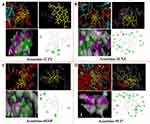
AFM technology is used to verify the binding of bioactive small molecules to proteins from the perspective of imaging, which offers intuitive image results of intermolecular interactions. In the AFM assay, the aggregation of proteins can prove that bioactive molecules bind to proteins. Bioactive molecules can affect the activity and function of proteins by changing their structure and state. The 2D configuration analysis in the AFM assay showed that the cPLA2 protein adsorbed on the quartz surface was a chain-like structure and was relatively dispersed before binding with aconitine (Figure 6A). The cPLA2 protein was aggregated and expanded violently after binding with aconitine under the same scanning conditions, and there were cPLA2 protein clumps on the quartz surface. Meanwhile, the length of the chain structure formed between cPLA2 proteins was shortened (Figure 6B). As shown in the 3D image, the surface of the cPLA2 protein is large, rough, loose, and dispersed, whereas cPLA2 became compact, smooth, and aggregated after binding with aconitine. When aconitine was bound to the protein, the degree of protein aggregation and height changed significantly; the latter reached approximately 1.1 times the initial height. AFM imaging confirmed that aconitine could bind to cPLA2 protein well, indicating that cPLA2 might be a potential direct target of aconitine. The combination of aconitine and cPLA2 may affect the function of proteins, leading to toxic effects.
 |
Figure 6 AFM analysis. AFM images (2D and 3D) of (A) cPLA2 and (B) aconitine acting on the protein. |
Discussion
Members of the Aconitum species, such as Aconiti Lateralis Radix Praeparata, Aconite Radix, and Aconiti Kusnezoffii Folium, are widely used in the clinic. Aconitine, a major bioactive alkaloid of Aconitum roots, has analgesic, anti-inflammatory, and antitumour effects.4 Aconitine is also the main toxic component of Aconitum plants, potentially causing varying degrees of cardiac injury by affecting energy metabolism, causing apoptosis, affecting ion channels, and via other mechanisms. However, the direct target proteins of aconitine-induced cardiotoxicity remain unclear. Target proteins are the key active components of TCMs that exert curative effects in vivo. The clarification of direct targets (ie, molecular initiation events) is the key to explaining the mechanism of drug efficacy or toxicity. Hence, the present study aimed to develop a new strategy for the identification and validation of potential direct targets of aconitine-induced cardiotoxicity. Network pharmacology and the DARTS technology were used to explore the potential targets of aconitine-induced cardiotoxicity. The obtained targets were screened via network integration analysis using the STRING database. Molecular docking was used to explain the possible binding sites between aconitine and related direct target proteins. Further literature investigation and AFM imaging confirmed that cPLA2 is a potential direct target of aconitine-induced cardiotoxicity.
In this study, the potential targets of aconitine-induced cardiotoxicity were predicted via network pharmacology; 152 common targets involving 41 closely related pathways were identified. The results of the degree value of targets and pathway analysis showed that aconitine may exert toxic effects through the PI3K-Akt, mTOR, and MAPK signalling pathways and the metabolism of arachidonic acid. Furthermore, aconitine can promote the release of inflammatory factors and further accelerate the inflammatory reaction, ultimately leading to myocardial injury. The PI3K/Akt/mTOR signalling pathway is required for the prosurvival signalling cascade, which participates in and regulates the apoptosis of cardiomyocytes.40 Transforming growth factor-β1 (TGF-β1) regulates the growth, development, and apoptosis of cardiomyocytes.41 Fuzi extracts can upregulate the expression of PI3K, p-Akt, mTOR, and TGF-β, and induce inflammation and apoptosis in cardiomyocytes; this indicates that Fuzi extracts exert cardiotoxic effects by activating the PI3K/Akt/mTOR and TNF-β signalling pathways and thus inducing cardiomyocyte apoptosis.42 Furthermore, reducing the expression of PI3K/Akt and mTOR can ameliorate myocardial hypertrophy and fibrosis, thereby reducing cardiomyocyte apoptosis.43 Tumour necrosis factor α (TNF-α) is an inflammatory cytokine produced in acute inflammation, which is related to the intracellular signal transduction pathway and can cause cell necrosis or apoptosis.44 Peng et al investigated the effects of aconitine on inflammation, apoptosis, and viability of H9c2 cardiomyocytes, and they found that aconitine upregulates the expression of FADD, cleaved-caspase 3; and inflammatory factors TNF-α, caspase-1, and IL-1 β; and it activates the cytochrome C and NLRP3 inflammasome. The results suggested that aconitine induced cardiomyocyte injury by attenuating BNIP3-dependent mitophagy and the TNF-α‐NLRP3 signalling axis.45 In the inflammatory pathway, the metabolism of arachidonic acid is related to the occurrence and development of myocardial injury. Its metabolism in myocardial tissue is regulated by phospholipases such as cPLA2 and sPLA2.46,47 PTGS2, also known as cyclooxygenase, has a high degree value in the analysis of key targets; its expression increases when cells are stimulated by inflammation. The upregulation of COX-2 and synthesis of prostaglandins promote the release of MMPs and further regulate collagen metabolism and the degree of fibrosis in myocardial tissue.24
Target proteins are the origin and biological basis of drug efficacy and toxicity. Aconitine reaches the target organs and acts on direct targets, which is the key molecular initiation event of its toxic effect. New strategies have been applied to identify the target proteins of natural products without any chemical modification, such as DARTS, CETSA, stability of proteins from rates of oxidation (SPROX), and thermal proteome profiling (TPP). CETSA can be used to study the effects of small molecules on the thermal stability of proteins in whole-cell lysates, cells, and tissues. However, the heating treatment may affect the permeability of the cell membrane, resulting in false positives, and subsequently affect the screening of target proteins. The limitation of SPROX technology is that it can only detect the binding of proteins that contain methionine to small molecules. In addition, not all methionine residues show different oxidation rates; thus, the interaction between proteins and ligands cannot be fully confirmed. TPP is an advanced CETSA method, which can be used to identify proteins that exhibit ligand-induced thermal stability at higher temperatures. However, some proteins are broken down at low temperatures or dissolved at high temperatures, which cannot be reproduced. By comparison, the DARTS technology is simple to operate and does not require washing or setting a temperature gradient. It can compensate for the shortcomings of SPROX, CETSA, and TPP and can be successfully applied to target screening with low affinity; thus, it has become the most popular method for identifying direct targets.48 Hence, based on the toxic concentration of aconitine-induced cardiomyocyte damage, a novel proteomic approach based on DARTS combined with LC-MS/MS was utilised to fish the direct target proteins of aconitine from myocardial cells, and the mechanism of toxicity was explored. The proteins directly interacting with aconitine were enriched from the total proteins of H9c2 cardiomyocytes via a molecular target fishing technique, and 15 potential direct targets were identified using LC-MS/MS. The 15 direct targets and targets predicted via network pharmacology were integrated and analysed using the STRING database. Through network topology analysis of PPI and KEGG, four direct target proteins (cPLA2, GYS1, CTNNA1, and ORC5) were screened as interactive. The molecular docking results showed that aconitine was bound to the active cavity of proteins via van der Waals forces or hydrogen bonds, and cPLA2 was identified as a direct target of aconitine based on literature investigation and biological pathway analysis. Moreover, cPLA2 is the upstream signal protein of PTGS2, and aconitine is speculated to act directly on the target protein cPLA2, which mediates the release of arachidonic acid, promotes the expression of PTGS2/COX-2, and further accelerates the release of inflammatory factors, ultimately leading to myocardial injury and dysfunction. Meanwhile, the immune activity of COX is strengthened, and COX-2 and its metabolites increase when myocardial ischemia or infarction occurs.36,37 Moreover, aconitine can promote the expression of PGE2 via the PGE2/COX-2 signalling pathway, thereby promoting the release of inflammatory factors and causing myocardial injury.39 AFM imaging verified that aconitine can bind to cPLA2 well. Thus, we further confirmed that cPLA2 is a direct target of aconitine-induced cardiotoxicity. However, the mechanism of aconitine-induced cardiotoxicity through the direct targets of GYS1, CTNNA1, and ORC5 remains unclear and will become the focus of our later studies. We also will verify the mechanism of aconitine-induced cardiotoxicity in vivo in the succeeding study. The present study not only contributes to understanding the process of aconitine acting on direct targets but also has great scientific significance in clarifying the mechanism of aconitine-induced cardiotoxicity and revealing its toxic nature.
Conclusion
In this study, we established a new strategy for the rapid identification of the direct targets of aconitine based on the DARTS technology and network integration analysis. Moreover, molecular docking, literature investigation, and AFM imaging were used to screen and verify the key direct targets. Four potential direct targets (cPLA2, GYS1, CTNNA1, and ORC5) were discovered by integrating and analysing the results of the DARTS technology and network pharmacology using the STRING database. The results of molecular docking showed that aconitine could bind to the four proteins well. Further studies have shown that cPLA2 is directly related to aconitine-induced cardiotoxicity. Finally, AFM imaging was used to verify the binding effect of cPLA2 and aconitine, which proved that cPLA2 was the key direct target of aconitine. By exploring the initial event of aconitine toxicity, we comprehensively explained the toxicity mechanism. The established research strategy laid a foundation for the target fishing of other toxic drugs.
Acknowledgments
This work was supported by the Scientific Research Project of the Tianjin Education Commission (2019KJ074).
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Wei XY, Qiu ZD, Chen JL, et al. Research advancement in mechanisms of processing and compatibility for detoxication of Aconitums. China J Chin Mater Med. 2019;44:3695–3704. doi:10.19540/j.cnki.cjcmm.20190629.301
2. Chinese Pharmacopoeia Commission. Chinese pharmacopeia. China Med Sci Press. 2020;Part I:40, 200, 247.
3. Bisset NG. Arrow poisons in China. Part II. Aconitum–botany, chemistry, and pharmacology. J Ethnopharmacol. 1981;4:247–336. doi:10.1016/0378-8741(81)90001-5
4. Zhou GH, Tang LY, Zhou XD, et al. A review on phytochemistry and pharmacological activities of the processed lateral root of Aconitum carmichaelii Debeaux. J Ethnopharmacol. 2015;160:173–193. doi:10.1016/j.jep.2014.11.043
5. Chan TYK. Aconite poisoning. Clin Toxicol. 2009;47:279–285. doi:10.1080/15563650902904407
6. Tian Z, Ma PY, Yang CY, et al. Study on arrhythmia induced by aconitine in rats. Chin J Labora Diagno. 2016;20:1447–1448.
7. Zhou SS, Yang J, Li YQ, et al. Effect of Cl−channel blockers on aconitine-induced arrhythmias in rat heart. Exp Physiol. 2005;90:865–872. doi:10.1016/j.nimb.2005.08.156
8. Liu F, Han X, Li N, et al. Aconitum alkaloids induce cardiotoxicity and apoptosis in embryonic zebrafish by influencing the expression of cardiovascular relative genes. Toxicol Lett. 2019;305:10–18. doi:10.1016/j.toxlet.2019.01.002
9. Zhang F, Cai L, Zhang J, et al. Aconitine-induced cardiac arrhythmia in human induced pluripotent stem cell-derived cardiomyocytes. Exp Ther Med. 2018;16:3497–3503. doi:10.3892/etm.2018.6644
10. Zhang X, Zhao BX, Song YQ, et al. Compatibility effect of aconitine and ginsenosides Rb1 on energy metabolism of primary cultured myocardial cells. Moderniza Tradit Chin Med Mater Med World Sci Technol. 2015;17:1785–1789. doi:10.11842/wst.2015.09.006
11. Zhao YN, Xie WD, Xing DM, et al. Advances in research approaches of action targets of active ingredients from Chinese herbs. Moderniza Tradit Chin Med Mater Med World Sci Technol. 2016;18:1005–1011. doi:10.11842/wst.2016.06.012
12. Song XQ, Zhang Y, Dai EQ, et al. Prediction of triptolide targets in rheumatoid arthritis using network pharmacology and molecular docking. Int Immunopharmacol. 2020;80:106179. doi:10.1016/j.intimp.2019.106179
13. Xie J, Gao S, Li L, et al. Research progress and application strategy on network pharmacology in Chinese materia medica. Chin Tradit Herbal Drugs. 2019;50:2257–2265. doi:10.7501/j.issn.0253-2670.2019.10.001
14. Wang X, Wang ZY, Zheng JH, et al. TCM network pharmacology: a new trend towards combining computational, experimental and clinical approaches. Chin J Nat Med. 2021;19:1–11. doi:10.1016/S1875-5364(21)60001-8
15. Wang S, Tian Y, Wang M, et al. Advanced activity-based protein profiling application strategies for drug development. Front Pharmacol. 2018;9:353. doi:10.3389/fphar.2018.00353
16. Pai MY, Lomenick B, Hwang H, et al. Drug affinity responsive target stability (DARTS) for small-molecule target identification. Methods Mol Biol. 2015;1263:287–298. doi:10.1007/978-1-4939-2269-7_22
17. Park YD, Sun W, Salas A, et al. Identification of multiple cryptococcal fungicidal drug targets by combined gene dosing and drug affinity responsive target stability screening. mBio. 2016;7:e1016–e1073. doi:10.1128/mBio.01073-16
18. Lu S, Tian Y, Luo Y, et al. Iminostilbene, a novel small-molecule modulator of PKM2, suppresses macrophage inflammation in myocardial ischemia-reperfusion injury. J Adv Res. 2021;29:83–94. doi:10.1016/j.jare.2020.09.001
19. Sun GJ, Li XM, Wei JX, et al. Pharmacodynamic substances in Salvia miltiorrhiza for prevention and treatment of hyperlipidemia and coronary heart disease based on lipidomics technology and network pharmacology analysis. Biomed Pharmacother. 2021;141:111846. doi:10.1016/j.biopha.2021.111846
20. Lomenick B, Hao R, Jonai N, et al. Target identification using drug affinity responsive target stability (DARTS). P Natl Acad Sci USA. 2009;106:21984–21989. doi:10.1073/pnas.0910040106
21. Gong F, Peng X, Sang Y, et al. Dichloroacetate induces protective autophagy in LoVo cells: involvement of cathepsin D/thioredoxin-like protein 1 and Akt-m TOR-mediated signaling. Cell Death Dis. 2013;4:e913. doi:10.1038/cddis.2013.438
22. Fleta-Soriano E, Martinez JP, Hinkelmann B, et al. The myxobacterial metabolite ratjadone A inhibits HIV infection by blocking the Rev/CRM1-mediated nuclear export pathway. Microb Cell Fact. 2014;13:17. doi:10.1186/1475-2859-13-17
23. Chin RM, Fu XD, Pai MY, et al. The metabolite α-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510:397–401. doi:10.1038/nature13264
24. Wang J, Lu LH, Wang Y, et al. Qishenyiqi Dropping Pill attenuates myocardial fibrosis in rats by inhibiting RAAS-mediated arachidonic acid inflammation. J Ethnopharmacol. 2015;176:375–384. doi:10.1016/j.jep.2015.11.023
25. Liang QR, Liu MJ, Hu BW. The histo- and cyto-chemical demonstration of NADH dehydrogenase in myocardium of aconitine poisoning rat. Chin J Foren Med. 1991;6:84–86. doi:10.13618/j.issn.1001-5728.1991.02.009
26. Pang YX, Nie QH, Liu XN, et al. Analysis of three cell lysates on total protein extraction in Western blot. J Hebei Med Univ. 2019;40:263–267. doi:10.3969/j.issn.1007-3205.2019.03.004
27. Yu HX, Li CY, Wang X, et al. Techniques and strategies for potential protein target discovery and active pharmaceutical molecule screening in a pandemic. J Proteome Res. 2020;19:4242–4258. doi:10.1021/acs.jproteome.0c00372
28. Shi MH, Wang JN, Xiao YJ, et al. Glycogen metabolism and rheumatoid arthritis: the role of glycogen synthase 1 in regulation of synovial inflammation via blocking AMP-activated protein kinase activation. Front Immunol. 2018;9:1714. doi:10.3389/fimmu.2018.01714
29. Pederson BA, Chen HY, Schroeder JM, et al. Abnormal cardiac development in the absence of heart glycogen. Mol Cell Biol. 2004;24:7179–7187. doi:10.1128/MCB.24.16.7179-7187.2004
30. Pan XQ, Yan BH, Zhou J, et al. Research progress of Chinese materia medica-induced cardiotoxicity. Prog Pharm Sci. 2020;44:730–742.
31. Dissanayake WC, Sorrenson B, Lee KL, et al. α-catenin isoforms are regulated by glucose and involved in regulating insulin secretion in rat clonal β-cell models. Biochem J. 2020;477:763–772. doi:10.1042/BCJ20190832
32. Wang XK, Wang QQ, Huang JL, et al. Novel candidate biomarkers of origin recognition complex 1, 5 and 6 for survival surveillance in patients with hepatocellular carcinoma. J Cancer. 2020;11:1869–1882. doi:10.7150/jca.39163
33. Yan H, Zhang CB, Li CH, et al. Progress in phospholipase A2 and its relevant Chinese materia medica. Moderniza Tradit Chin Med Mater Med World Sci Technol. 2013;15:1620–1629. doi:10.11842/wst.2013.07.025
34. McHowat J, Creer MH. Catalytic features, regulation and function of myocardial phospholipase A2. Curr Med Chem Cardiovasc Hematol Agents. 2004;2:209–218. doi:10.2174/1568016043356282
35. Pietruszewska W, Fendler W, Podwysocka M, et al. Expression of transcript variants of PTGS1 and PTGS2 genes among patients with chronic rhinosinusitis with nasal polyps. Diagnostics (Basel). 2021;11:135. doi:10.3390/diagnostics11010135
36. Bolli R, Shinmura K, Tang XL, et al. Discovery of a new function of cyclooxygenase (COX)-2: COX-2 is a cardioprotective protein that alleviates ischemia/reperfusion injury and mediates the late phase of preconditioning. Cardiovasc Res. 2002;55:506–519. doi:10.1016/S0008-6363(02)00414-5
37. Saito T, Rodger IW, Hu F, et al. Inhibition of COX pathway in experimental myocardial infarction. J Mol Cell Cardiol. 2004;37:71–77. doi:10.1016/j.yjmcc.2004.04.002
38. Cheng I, Liu X, Plummer SJ, et al. COX2 genetic variation, NSAIDs, and advanced prostate cancer risk. Brit J Cancer. 2007;97:557–561. doi:10.1038/sj.bjc.6603874
39. Cheng C, Zeng YL, Wang J, et al. Effect observation of aconitine on the expression of PGE2 in peripheral blood mononuclear cells of normal 615 mice and its regulation on treg. J Liaoning Univ Tradit Chin Med. 2020;22:42–46. doi:10.13194/j.issn.1673-842x.2020.04.011
40. Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114:549–564. doi:10.1161/CIRCRESAHA.114.302022
41. Jian B, Xu J, Connolly J, et al. Serotonin mechanisms in heart valve disease I serotonin-induced up-regulation of transforming growth factor- beta 1 via G-protein signal transduction in aortic valve interstitial cells. Am J Pathol. 2002;161:2111–2121. doi:10.1016/S0002-9440(10)64489-6
42. Huang GY, Yang L, Zhou W, et al. Study on cardiotoxicity and mechanism of “fuzi” extracts based on metabonomics. Int J Mol Sci. 2018;19:3506. doi:10.3390/ijms19113506
43. Yu YH, Hu ZY, Li B, et al. Ivabradine improved left ventricular function and pressure overload-induced cardiomyocyte apoptosis in a transverse aortic constriction mouse model. Mol Cell Biochem. 2019;450:25–34. doi:10.1007/s11010-018-3369-x
44. Idriss HT, Naismith JH. TNF alpha and the TNF receptor superfamily: structure-function relationship(s). Microsc Res Techniq. 2000;50:184–195. doi:10.1002/1097-0029(20000801)50:33.0.CO;2-H
45. Peng F, Zhang N, Wang CT, et al. Aconitine induces cardiomyocyte damage by mitigating BNIP3‐dependent mitophagy and the TNFα‐NLRP3 signalling axis. Cell Proliferat. 2020;53:e12701. doi:10.1111/cpr.12701
46. Dong LY, Chen ZW. Myocardial ischemia/reperfusion injury and inflammatory reaction. Chin J Clin Pharm Th. 2008;13:582–588.
47. Yuan L, Zhou LJ. Effect of arachidonic acid metabolism on cardiac fibrosis associated with inflammation. Adv Cardiovasc Dis. 2010;31:62–65. doi:10.3969/j.issn.1004-3934.2010.01.020
48. Chang J, Kim Y, Kwon HJ. Advances in identification and validation of protein targets of natural products without chemical modification. Nat Prod Rep. 2016;33(5):719–730. doi:10.1039/c5np00107b
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.