Back to Journals » OncoTargets and Therapy » Volume 9
A negative regulation loop of long noncoding RNA HOTAIR and p53 in non-small-cell lung cancer
Authors Zhai N, Xia Y, Yin R, Liu J, Gao F
Received 9 April 2016
Accepted for publication 15 June 2016
Published 16 September 2016 Volume 2016:9 Pages 5713—5720
DOI https://doi.org/10.2147/OTT.S110219
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Nailiang Zhai,1 Yongfu Xia,1 Rui Yin,2 Jinping Liu,3 Fuquan Gao1
1Department of Respiratory Medicine, Affiliated Hospital of Binzhou Medical University, 2Department of Respiratory Medicine, People’s Hospital of Binzhou City, 3Department of Pharmacology, Binzhou Medical University, Binzhou, Shandong, People’s Republic of China
Abstract: Non-small-cell lung cancer (NSCLC) is one of the leading causes of cancer-related death worldwide, and the 5-year survival rate is still low despite advances in diagnosis and therapeutics. A long noncoding RNA (lncRNA) HOX antisense intergenic RNA (HOTAIR) has been revealed to play important roles in NSCLC carcinogenesis but the detailed mechanisms are still unclear. In the current study, we aimed to investigate the regulation between the lncRNA HOTAIR and p53 in the NSCLC patient samples and cell lines. Our results showed that HOTAIR expression was significantly higher in the cancer tissues than that in the adjacent normal tissue, and was negatively correlated with p53 functionality rather than expression. When p53 was overexpressed in A549 cells, the lncRNA HOTAIR expression was downregulated, and the cell proliferation rate and cell invasion capacity decreased as a consequence. We identified two binding sites of p53 on the promoter region of HOTAIR, where the p53 protein would bind to and suppress the HOTAIR mRNA transcription. Inversely, overexpression of lncRNA HOTAIR inhibited the expression of p53 in A549 cells. Mechanistic studies revealed that HOTAIR modified the promoter of p53 and enhanced histone H3 lysine 27 trimethylation (H3K27me3). These studies identified a specific negative regulation loop of lncRNA HOTAIR and p53 in NSCLC cells, which revealed a new understanding of tumorigenesis in p53 dysfunction NSCLC cells.
Keywords: NSCLC, LncRNA HOTAIR, p53, negative loop
Introduction
Lung cancer is the second common malignant tumors and the leading cause of cancer death in the US and Asia.1,2 Lung cancer is characterized by high mortality and low cure rates, with an average of 5-year survival rate <18% based on SEER Cancer Statistics Review 1975–2012 conducted by the National Cancer Institute in the US.3 Among all the lung cancer incidents, non-small-cell lung cancer (NSCLC) accounts for ~85%.4,5 The treatment of NSCLC is mainly surgery, and radio- or chemotherapy; specific treatment varies from case to case depending on the stage of the tumor. Up to now, a number of site-specific targeting chemotherapeutic drugs have been used in treating NSCLC patients especially those in metastatic stages and have been received promising results in clinic.6–8 In all these therapies, the principle strategy is to induce DNA damage. As p53 mediates cellular response to DNA damage, the study of the roles of p53 in NSCLC becomes obviously essential.
P53 functions in response to a variety of cellular stress signals to induce cell cycle arrest, apoptosis, or senescence,9,10 which led to reduced cell proliferation rate and cell death.11 One of p53 downstream effectors, p21, is a potent cyclin-dependent kinase inhibitor. When activated, p21 inhibits CDK1, CKD2, and CDK4/6 complex, resulting in cell cycle arrest at G1 and S phases.12 New clinical research indicates that the activation of p53/p21 is associated with poor prognosis in NSCLC patients.13 The disruption of p53 pathway is a well-known mechanism to drive tumor formation, which was discovered in many types of cancers. The research by Mogi and Kuwano showed that P53 gene is mutated in 50% of all human cancers and >70% of lung cancer.14 These cancer-driving mutations on P53 gene cause the loss of the tumor suppressing function of p53. A recent study has shown that a longtime inactivation of P53 gene is necessary for significant tumor growth, but the restoration of wide type p53 can stop this growth and trigger tumor elimination.15 However, the mechanisms revealing p53 dysfunction are still obscure so far.
Noncoding RNAs fulfill a diversity of biological functions, including gene regulation, RNA splicing, DNA synthesis, genome defense, and so on. They can be divided into small noncoding RNAs (<200 kb) and long noncoding RNAs (lncRNAs) (>200 kb) based on their length. Since the discovery, lncRNAs were found as the cause for a variety of diseases, such as cancers, neurological disorders, and fragile X syndrome. Recent evidence has shown that many lncRNAs are heavily involved in both tumor oncogenic and suppressive pathways;16–18 therefore, they have gained increasing attention in their gene regulatory functions. HOX antisense intergenic RNA (HOTAIR) is a lncRNA that locates on human chromosome 12 loci 13.19 Discovered by Howard Chang’s group, HOTAIR was shown to recruit polycomb repressive complex 2 (PRC2), a transcriptional co-repressor, to decrease the transcription of the homeobox gene D cluster and some other genes.19,20 This is an example of gene silencing at long distance, which has been used frequently in development and disease initiation. HOTAIR’s role in cancer has been shown by several studies. Its expression is significantly higher in hepatocellular carcinoma compared to adjacent noncancerous tissues.21 A similar role of HOTAIR was also found in pancreatic cancer.22 HOTAIR was also reported to facilitate breast cancer metastasis and loss of HOTAIR reduced cancer invasiveness.23 A number of lncRNAs have been found under direct p53 regulation or shown correlation with p53 level;19,24 yet, the correlation between HOTAIR and p53 has not been studied.
In this study, we intended to determine whether there was a correlation between HOTAIR and P53 in NSCLC patients and cell lines. Furthermore, the effect of cell proliferation rate and invasion capability was measured in p53 overexpressed NSCLC cell lines in which HOTAIR was dysregulated. Finally, our bioinformatics analysis predicted that p53 regulates HOTAIR by direct interaction, which was confirmed by experiments.
Methods
Patient specimens and cell culture
A total of 48 NSCLC tissues were obtained from NSCLC patients receiving surgical treatment at the Department of Respiration Medicine, Affiliated Hospital of Binzhou Medical University between January 2014 and February 2015. All patients recruited in this study were not treated before they diagnosed with NSCLC based on histopathological evaluation. All collected tissue samples were immediately stored at liquid nitrogen until use. Informed consent was obtained from all patients with regard to surgical treatment and acquisition of tissue specimens. The study was approved by the Ethics Committee of the Binzhou Medical University. Human tissue acquisition and use in this study complied with the National Regulations on the Use of Clinical Samples in China. The NSCLC cell lines A549 (P53 wild-type), NCI-H157 (P53 mutant), and H1299 (P53 deletion) were obtained from Cellbio Company (Shanghai, People’s Republic of China). These cell lines were cultured at 37°C in a CO2 incubator in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin.
The pcDNA3-flag-p53 (#10838) plasmid and the lncRNA HOTAIR expressing plasmid LZRS-HOTAIR (#26110) were purchased from Addgene (Cambridge, MA, USA); the pcDNA3-flag vector control and pGL3-basic vector were gifts from Dr Zhiqiang Liu, Department of Pathophysiology, Tianjin Medical University. The promoter of lncRNA HOTAIR was cloned into the pGL3-basic firefly luciferase reporter with the primers 5′-ACTGGTACCTAAGCGGAGAGAGTCCC-3′ and 5′-ACTAAGCTTGAGTCAGAGTTCCCCAC-3′. Plasmid transfection was achieved by Lipofectamine 2000 reagent (Life Technology, Shanghai, People’s Republic of China) with ratio of Lipo: DNA =2; after 6 hours, the media were changed to complete culture media for another 48 hours.
Real-time PCR analysis of mRNA expression levels
Total RNA was extracted by Trizol reagent (Invitrogen, Carlsbad, CA, USA). Quantitative polymerase chain reaction (PCR) was performed using specific primers (Sangon, Shanghai, People’s Republic of China) for HOTAIR. The cycling conditions were 30 seconds polymerase activation at 95°C, followed by 40 cycles amplification at 95°C for 5 seconds, and extension at 60°C for 30 seconds. The first strand cDNA served as the template for the regular PCR performed using a DNA Engine (Applied Biosystem, CA, USA). PCR primers: HOTAIR (200 bp), forward 5′-GGCAAATGTCAGAGGGTTCT-3′ and reverse 5′-TTCTTAAATTGGGCTGGGTC-3′; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (173 bp), forward 5′-ACAGGGGAGGTGATAGCATT-3′ and reverse 5′-GACCAAAAGCCTTCATACATCTC-3′. GAPDH as an internal control was used to normalize the data to determine the relative abundance of the target RNA. The amplification curves and melting curves were analyzed after completion of the reaction. Gene expression values were represented using the 2−ΔΔCt method.
Western blot
Cells were lysed by radioimmunoprecipitation buffer and proteins were then extracted and quantified by Bradford reagent. An amount of 50 mg of total proteins were loaded on Nupage 4%–12% polyacrylamide gels (Thermo Fisher Scientific, Waltham, MA, USA) for each sample and transferred to polyvinylidene difluoride membranes (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA), then they were probed with the following antibodies separately: mouse anti-p53 (1:1,000, Cat No: ab28, Abcam, Cambridge, UK) and rabbit anti-β-actin (1:2,000, Cat No: ab3280, Abcam). For detection, goat antirabbit or goat antimouse secondary antibodies conjugated to horseradish peroxidase (1:2,000, GE Healthcare Bio-Sciences Corp.) were used. Protein abundance was measured by chemiluminescence reaction (ECL, GE Healthcare Bio-Sciences Corp.). Western blot and quantification were performed using ImageJ software analysis (National Institutes of Health, Bethesda, MD, USA).
Cell proliferation assays
Cell Proliferation Reagent Kit I (MTT) (Roche Applied Science, IN, USA) was used to monitor cell proliferation rate. A total of 3,000 cells were seeded into each well of a 96-well plate. All experiments were performed in triplicate and cell proliferation rates were recorded every 24 hours according to the manufacturer’s protocol.
In vitro invasion assay
A modified in vitro Boyden chamber invasion assay with Matrigel-coated Transwell chambers was performed. Cells were trypsinized and resuspended in serum-free DMEM medium, and then were added to rehydrated Matrigel-coated inserts (BioCoat Matrigel Invasion Chamber; NJ, USA); the inserts were then placed in 24-well companion trans plates with DMEM medium and 10% fetal bovine serum (chemoattractant) for cell migration through the pores to the trans side. After 24 hours, the cells and matrigel in the upper chambers were removed, and the cells in the bottom trans chambers were fixed with 3% glutaraldehyde and stained with 4′,6-diamidino-2-phenylindole (DAPI) (1:5,000; Sigma-Aldrich Co., St Louis, MO, USA) fluorescent for 15 minutes. DAPI-stained cells were counted in five randomly different fields under an inverted fluorescence microscope. The experiments were performed in triplicate wells and each experiment was performed in triplicate.
Chromatin immunoprecipitation assay
Cells were cross-linked in 1% (vol/vol) formaldehyde for 10 minutes at room temperature. Then, 0.125 M glycine was used to quench the cross-linking reaction. Cells were pelleted and washed with ice-cold phosphate-buffered saline. Next, cell nuclei were isolated and a Bioruptor sonicator (Diagenode) was used to shear chromatin DNA. An amount of 5 μL of p53 antibody (1:2,000, Cat No: ab28, Abcam) diluted in phosphate-buffered saline buffer was added to Dynabeads Protein A (Invitrogen) beads and incubated for 3 hours at 4°C with rotation. An amount of 5 μL of normal rabbit immunoglobulin G (Cell Signaling, CA, USA) was used as negative control. Then, the dynabeads-antibody complexes were incubated with sheared chromatin DNA overnight at 4°C. After immunoprecipitation, the precipitated complex was treated with RNase A and Proteinase K, and incubated at 65°C overnight to reverse cross-links. Primers were designed and quantitative RT-PCR (qRT-PCR) was performed. The sequence of primers for HOTAIR promoter (110 bp) was as follows: F 5′-GCCCTTCTCCTAGCCCACCG-3′, R 5′-GTGGGGACCCGCTAGACCTG-3′; sequence of primers for p53 promoter (125 bp) was as follows: F 5′-CAAAGAAATGGAGCCGTGTA-3′, R 5′-GCCTCCTAAAGTGCCAAGAT-3′. Chromatin immunoprecipitation (ChIP)-qPCR data were normalized either by the percent input method or relative to the immunoglobulin G control.
Statistical analysis
Statistical analysis was performed using the SPSS statistical software program Version 15.0 (SPSS Inc., Chicago, IL, USA). Data from all these experiments were reported as mean ± standard deviation for each group. Intergroup differences were analyzed by one-way analysis of variance. Correlation between expressions of p53 and lncRNA HOTAIR was analyzed with regression analysis. Differences with P-value of <0.05 were considered statistically significant.
Results
The lncRNA HOTAIR expression was negatively correlated with p53 in NSCLC
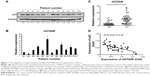
First of all, we measured the expression of p53 via real-time PCR in all 48 NSCLC patients’ samples, and found that the expression was differed from patient to patient (data not shown). The expression of p53 in 12 samples from all 48 patients was also measured by Western blot (Figure 1A), and mRNA level of HOTAIR was measured by qRT-PCR as well (Figure 1B). In addition, expressions of the lncRNA HOTAIR in the cancer tissues or the adjacent-normal tissues from all 48 samples were detected using real-time PCR, and the result showed that expression of HOTAIR in cancer tissue group was significantly higher than that in the adjacent-normal tissue control group (P<0.001) (Figure 1C). By comparing the expression of p53 and HOTAIR, we found that the higher level of HOTAIR was associated with a lower p53 level. The same negative correlation between lncRNA HOTAIR and P53 was confirmed in all of our 48 NSCLC patients’ samples using regression analysis (Figure 1D).
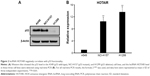
In order to further investigate the correlation between the expressions of HOTAIR and p53, the expressions of both molecules were measured in three NSCLC cell lines based on their p53 function and presence: A549 (wild-type p53), NCI-H157 (mutant p53), and H1299 (p53 deletion). We confirmed the expression characteristics of p53 in these three cell lines, among which H1299 cells had almost invisible p53, while the A549 and NCI-H157 cells had considerable p53 expression (Figure 2A). However, we did not detect the mutation of p53 in NCI-H157 cells in this study. Next, the expression level of HOTAIR was measured in each of the three cell lines by qRT-PCR. We found that HOTAIR level was significantly higher in H1299 cells and NCI-H157 cells compared to A549 cells (Figure 2B), which suggested that the expression levels of HOTAIR were negatively correlated with p53 functionality rather than expression.
Modulation of p53 expression alternated the lncRNA HOTAIR expression and cell invasion in NSCLC cells
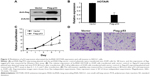
As we observed the negative correlation between p53 and HOTAIR, we next investigated if the expression of p53 suppressed HOTAIR level. p53 was overexpressed in A549 cells using the pcDNA3-flag-p53 plasmid, and Western blot was used to confirm the high protein level of p53 48 hours after transfection (Figure 3A). Then, the expression of HOTAIR was measured by qRT-PCR, and our result showed that the expression of HOTAIR was significantly decreased in the p53 overexpressing cells compared to the cells transfected with vector control (Figure 3B). Next, phenotypes changed in cell biology of the A549 cells were examined. We confirmed that p53 overexpression affected cell proliferation and migration in the A549 NSCLC cells, as overexpression of p53 significantly decreased the proliferation rate of A549 cells (Figure 3C) as well as the invasion capability in vitro (Figure 3D), compared to the cells transfected with vector control, respectively.
p53 Could bind and regulate the HOTAIR promoter
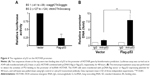
As the negative correlation between p53 and HOTAIR was known, we next studied possible interaction between these two molecules via bioinformatics analysis. Based on our computational prediction, two p53 binding sites were found on the promoter region of HOTAIR (Figure 4A, taller). In order to verify the prediction, the HOTAIR promoter was cloned into the pGL3-luciferase plasmid to construct HOTAIR-luc, which fused luciferase to HOTAIR promoter. Next, the HOTAIR-luc and p53 plasmids were cotransfected into A549 cells, in which the expression of p53 was then confirmed (data not shown). We found that the luciferase activity of the HOTAIR promoter was significantly decreased in cells overexpressing p53 compared with the vector control (Figure 4A, shorter). In order to further elaborate how P53 regulates HOTAIR expression at transcriptional level, ChIP assay was carried out in A549 cells. We found the HOTAIR promoter was pulled down significantly more in the p53 overexpressing cells than that in cells transfected with empty vector (Figure 4B), suggesting p53 binds and regulates the lncRNA HOTAIR promoter.
lncRNA HOTAIR reversely regulated the histone H3 lysine 27 trimethylation of p53 gene promoter
HOTAIR interacts with and recruits PRC2 to regulate the chromosome occupancy through the EZH2 molecule (a subunit of PRC2), which leads to histone H3 lysine 27 trimethylation (H3K27me3) of some genes.25 Based on this known information, we explored whether there is a negative loop between p53 and HOTAIR. When the HOTAIR overexpression plasmid was transiently transfected into A549 cells for 48 hours, the protein level of p53 was obviously downregulated when detected by Western blot assay (Figure 5A), and we found that P53 mRNA level was also significantly downregulated shown by real-time PCR assay (Figure 5B). In addition, the epigenetic modification of p53 promoter was checked via ChIP assay, which showed that overexpression of HOTAIR could increase the H3K27me3 level (Figure 5C), whereas the H3K4me3 was shown slightly decreased in both cells (data not shown). This result implies that HOTAIR might epigenetically regulate the P53 gene promoter in a negative feedback loop model.
Discussion
Lung carcinoma is one of the most common types of cancer worldwide and exists as a major concern of health problem. As the most popular subtype of lung cancer, NSCLC occurred >50% in Asian female lung cancer patients. Although significant progress has been made in the past decades in chemotherapy, NSCLC is still resistant to current treatments. Therefore, better understanding of the mechanisms leading to NSCLC formation and development is essential. In this study, we found that p53 is involved in NSCLC cell proliferation and migration, and a negative correlation was identified between p53 functionality and an lncRNA molecule, HOTAIR. Further analysis discovered that p53 represses HOTAIR expression by direct binding to its promoter. Inversely, lncRNA HOTAIR inhibited the expression of p53 in NSCLC cells, and mechanistic studies revealed that HOTAIR modified the promoter of p53 and enhanced histone H3 lysine 27 trimethylation (H3K27me3). Therefore, we for the first time revealed a negative regulation loop between p53 and lncRNA HOTAIR in NSCLC.
lncRNAs have attracted increasing attentions in recent years as they appeared to be involved in a number of cancer formations and metastases. Many lncRNAs have shown the capacity to regulate the initiation, progression, and response to therapy in a variety of solid tumors and hematological malignancies.26,27 Gene expression microarrays identified that hundreds of lncRNAs are associated with lung cancer and massively parallel RNA sequencing of tumor tissues and paired adjacent nontumor tissues in the lung.28–31 Thus, further study on the role of lncRNAs in NSCLC is worth conducting. Recent research identified HOTAIR as a promising target to diagnose and treat lung cancer. A study discovered that NSCLC tumor samples had higher HOTAIR expression levels compared to their corresponding noncancerous lung tissue.32 Besides lung cancer and breast cancer,23 the expression levels of HOTAIR were found elevated in several other types of tumor tissues as well, including liver cancer33 and certain colorectal cancer.34 Colorectal cancer patients who showed a high HOTAIR expression level also had a worse prognosis than those patients with low HOTAIR levels. Besides primary tumor, HOTAIR is also found involved in tumor metastasis, for example, lymph node metastasis in hepatocellular carcinoma.21 Therefore, HOTAIR could be a promising diagnostic target, which needs further confirmation.
Here, we discovered that p53 was involved in the regulation of HOTAIR expression in NSCLC cell lines by direct binding to its promoter. Our study identified a new p53 downstream regulatory mechanism that led to cancer when nonfunctional. P53 is one of the most powerful tumor suppressor genes identified in human cells. Its mutation is most frequently found in cancer cells and particularly lung cancer, and >70% of lung cancer cases have a P53 mutation. As HOTAIR is under the regulation of p53, deregulation of HOTAIR expression level in lung cancer could be common as well. Therefore, potential therapeutic drugs targeting this lncRNA might be able to provide promising results.
The function of HOTAIR in NSCLC is still not clear and needs further investigation. However, a previous study on breast cancer found that HOTAIR promotes primary cancer formation and metastasis by recruiting PRC2, which methylate H3K27 and silence metastasis suppressor genes.23 Furthermore, HOTAIR can serve as a scaffold for multiple histone modification complexes: the 5′ end of HOTAIR binds with PRC2, and its 3′ end binds to lysine (K)-specific demethylase 1A complex.35 Therefore, this bifunctional character of HOTAIR could be required for coordinating histone modifications of H3K27 methylation and H3K4 demethylation for epigenetic gene silencing in the metastatic processes.36 In the current study, the relationship between HOTAIR and p53 has been verified. We found that the H3K27me3 modification on the promoter of p53 was changed, which might be relative with HOTAIR. Potential research might focus on the negative feedback loop between p53 and HOTAIR.
Conclusion
In conclusion, our study demonstrated that the p53 levels was negatively correlated with the amount of HOTAIR in NSCLC patients’ samples and cell lines, suggesting a regulatory role of p53 in HOTAIR transcription. Moreover, the HOTAIR might regulate the expression of p53 epigenetically via changing the modification of H3K27me3 with a negative feedback loop. This work added another piece of evidence in HOTAIR’s role in cancer promotion and metastasis, and highlighted the importance of HOTAIR as a potential target in cancer diagnostic and treatment.
Disclosure
The authors report no conflicts of interest in this work.
References
Saika K, Machii R. Cancer mortality attributable to tobacco in Asia based on the WHO global report. Jpn J Clin Oncol. 2012;42(10):985. | ||
Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212–236. | ||
Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–241. | ||
Li C, Zhang S, Lu Y, Zhang Y, Wang E, Cui Z. The roles of Notch3 on the cell proliferation and apoptosis induced by CHIR99021 in NSCLC cell lines: a functional link between Wnt and Notch signaling pathways. PLoS One. 2013;8(12):e84659. | ||
Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011;32(4):605–644. | ||
Soria JC, Mauguen A, Reck M, et al. Systematic review and meta-analysis of randomised, phase II/III trials adding bevacizumab to platinum-based chemotherapy as first-line treatment in patients with advanced non-small-cell lung cancer. Ann Oncol. 2013;24(1):20–30. | ||
Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. | ||
Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371(21):1963–1971. | ||
Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. | ||
Wu L, Levine AJ. Differential regulation of the p21/WAF-1 and mdm2 genes after high-dose UV irradiation: p53-dependent and p53-independent regulation of the mdm2 gene. Mol Med. 1997;3(7):441–451. | ||
Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22(56):9030–9040. | ||
Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65(10):3980–3985. | ||
Xie D, Lan L, Huang K, et al. Association of p53/p21 expression and cigarette smoking with tumor progression and poor prognosis in non-small cell lung cancer patients. Oncol Rep. 2014;32(6):2517–2526. | ||
Mogi A, Kuwano H. TP53 mutations in nonsmall cell lung cancer. J Biomed Biotechnol. 2011;2011:583929. | ||
Wiman KG. Pharmacological reactivation of mutant p53: from protein structure to the cancer patient. Oncogene. 2010;29(30):4245–4252. | ||
Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457(7228):413–420. | ||
Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21(3):416–425. | ||
Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2(11):986–991. | ||
Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. | ||
Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. | ||
Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39(6):2119–2128. | ||
Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. | ||
Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. | ||
Isin M, Ozgur E, Cetin G, et al. Investigation of circulating lncRNAs in B-cell neoplasms. Clin Chim Acta. 2014;431:255–259. | ||
Zhang J, Zhang P, Wang L, Piao HL, Ma L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin. 2014;46(1):1–5. | ||
Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. | ||
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. | ||
White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15(8):429. | ||
Yang J, Lin J, Liu T, et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer. 2014;85(2):110–115. | ||
Wang Y, Chen W, Chen J, Pan Q, Pan J. LncRNA expression profiles of EGFR exon 19 deletions in lung adenocarcinoma ascertained by using microarray analysis. Med Oncol. 2014;31(9):137. | ||
Xu G, Chen J, Pan Q, et al. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One. 2014;9(8):e104044. | ||
Nakagawa T, Endo H, Yokoyama M, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436(2):319–324. | ||
Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. | ||
Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35(7):1510–1515. | ||
Sun X, Du P, Yuan W, et al. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015;6:e1907. | ||
Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856(1):151–164. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.