Back to Journals » International Journal of Nanomedicine » Volume 18
A Multivalent and Thermostable Nanobody Neutralizing SARS-CoV-2 Omicron (B.1.1.529)
Authors Lu Y, Li Q, Fan H , Liao C, Zhang J, Hu H , Yi H, Peng Y, Lu J, Chen Z
Received 23 August 2022
Accepted for publication 20 December 2022
Published 19 January 2023 Volume 2023:18 Pages 353—367
DOI https://doi.org/10.2147/IJN.S387160
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Dongwoo Khang
Yuying Lu,1– 5 Qianlin Li,1– 5 Huahao Fan,6 Conghui Liao,1– 5 Jingsong Zhang,1– 5 Huan Hu,1– 5 Huaimin Yi,1– 5 Yuanli Peng,1– 5 Jiahai Lu,1– 5 Zeliang Chen1– 3,7
1One Health Center of Excellence for Research and Training, School of Public Health, Sun Yat-Sen University, Guangzhou, People’s Republic of China; 2National Medical Products Administration Key Laboratory for Quality Monitoring and Evaluation of Vaccines and Biological Products, Guangzhou, People’s Republic of China; 3Key Laboratory of Tropical Diseases Control, Sun Yat-Sen University, Ministry of Education, Guangzhou, People’s Republic of China; 4Research Institute of Sun Yat-Sen University in Shenzhen, Shenzhen, People’s Republic of China; 5Hainan Key Novel Thinktank “Hainan Medical University ‘One Health’ Research Center”, Haikou, People’s Republic of China; 6College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, People’s Republic of China; 7Key Laboratory of Zoonose Prevention and Control at Universities of Inner Mongolia Autonomous Region, Medical College, Inner Mongolia Minzu University, Tongliao, People’s Republic of China
Correspondence: Jiahai Lu; Zeliang Chen, One Health Center of Excellence for Research and Training, School of Public Health, Sun Yat-Sen University, Guangzhou, People’s Republic of China, Email [email protected]; [email protected]
Background: The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variants have risen to dominance, which contains far more mutations in the spike protein in comparison to previously reported variants, compromising the efficacy of most existing vaccines or therapeutic monoclonal antibodies. Nanobody screened from high-throughput naïve libraries is a potential candidate for developing preventive and therapeutic antibodies.
Methods: Four nanobodies specific to the SARS-CoV-2 wild-type receptor-binding domain (RBD) were screened from a naïve phage display library. Their affinity and neutralizing activity were evaluated by surface plasmon resonance assays, surrogate virus neutralization tests, and pseudovirus neutralization assays. Preliminary identification of the binding epitopes of nanobodies by peptide-based ELISA and competition assay. Then four multivalent nanobodies were engineered by attaching the monovalent nanobodies to an antibody-binding nanoplatform constructed based on the lumazine synthase protein cage nanoparticles isolated from the Aquifex aeolicus (AaLS). Finally, the differences in potency between the monovalent and multivalent nanobodies were compared using the same methods.
Results: Three of the four specific nanobodies could maintain substantial inhibitory activity against the Omicron (B.1.1.529), of them, B-B2 had the best neutralizing activity against the Omicron (B.1.1.529) pseudovirus (IC50 = 1.658 μg/mL). The antiviral ability of multivalent nanobody LS-B-B2 was improved in the Omicron (B.1.1.529) pseudovirus assays (IC50 = 0.653 μg/mL). The results of peptide-based ELISA indicated that LS-B-B2 might react with the linear epitopes in the SARS-CoV-2 RBD conserved regions, which would clarify the mechanisms for the maintenance of potent neutralization of Omicron (B.1.1.529) preliminary.
Conclusion: Our study indicated that the AaLS could be used as an antibody-binding nanoplatform to present nanobodies on its surface and improve the potency of nanobodies. The multivalent nanobody LS-B-B2 may serve as a potential agent for the neutralization of SARS-CoV-2 variants.
Keywords: COVID-19, bio-nanotechnology, self-assembly, nanobody multimers, thermal stability
Introduction
The global spread of coronavirus disease 2019 (COVID-19) has led to an unprecedented pandemic due to its high infectivity and its impact on human health and society. Given the increase in the number of infected individuals and the acceleration of the transmission speed, it provides sufficient natural selection opportunities for the virus, thus producing more mutations and providing a better breeding ground for virus mutation. The angiotensin-converting enzyme 2 (ACE2) receptor-binding domain (RBD) region of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein is the primary region of convergent mutations in circulating variants of SARS-CoV-2. Indeed, a variety of RBD-mutated variants appeared, which accelerated the spread of SARS-CoV-2 or allowed it to evade antibody neutralization, resulting in the decrease of antibody efficacy elicited by vaccines or the effect of protective neutralizing antibodies.1–5 The SARS-CoV-2 Omicron (B.1.1.529) variant which has an unprecedented number of mutations attracts worldwide attention, with 15 mutations in RBD, while the previous Beta (B.1.351) variant has 3 mutations (K417N, E484K, and N501Y) and Delta (B.1.617.2) variant has only 2 mutations in RBD (L452R and T478K).3,6 The amino acid site mutation of RBD, especially the receptor-binding motif (RBM), affects RBD interaction with ACE2, thus increasing the affinity of S protein with ACE2.4,7 Most neutralizing antibodies occupy the neck and shoulder epitopes of RBD, blocking the interaction between S protein and ACE2, thus preventing viruses from attaching to host cells.8 However, of the currently licensed or approved therapeutic monoclonal antibodies, only the antibody S309 and COV2-2196/COV2-2130 cocktails can neutralize the Omicron (B.1.1.529) variant, although the neutralization potency is also reduced by at least one-third.9 These antibodies, which retain neutralizing activity against Omicron (B.1.1.529) variant, can destabilize the S-trimer, and hardly block the interaction of ACE2 with RBD due to their antigen-binding sites being far away from the ACE2 binding site on the S protein.7,10
Therefore, it is urgent to develop a multivalent antibody against multiple epitopes and more conserved epitopes of SARS-CoV-2. It would be able to target the highly conserved epitopes on S protein and use antibodies that recognize different epitopes on S protein in combination covering the neutralizing epitopes on S protein or RBD as much as possible.10,11 At present, there are several reports that SARS-CoV-2-neutralizing single-domain antibodies from naïve phage display library or immunization have a neutralizing effect on SARS-CoV-2, which can interfere with the interaction between RBD and ACE2.12–14 Also, existing nanobodies binding to highly conserved epitopes on SARS-CoV-2 RBD can cross neutralize SARS-CoV.15 These nanobodies are considered potential therapeutics for COVID-19 patients.
Nanobodies are the variable regions of heavy chains of camelid antibodies and lack the first constant regions in conventional antibodies, so nanobodies are typically only 12–14 kDa in size, with dimensions in the nanometer range (about 2.5 nanometers in diameter and 4 nanometers in height).16,17 Due to the small size of the nanobodies, the complementarity determining region 3 (CDR3) is longer than that of humans, forming a finger-like structure, which can penetrate the cavity on the surface of the antigen, so that the nanobodies can bind to epitopes that traditional antibodies are not accessible.18 In addition to being the smallest unit that retains the antigenic affinity and specificity, nanobodies have an important advantage over traditional antibodies that they can be easily assembled into multimers. As monomers, especially nanobodies isolated from nonimmune synthetic libraries usually lack the high binding affinity required for therapeutic use or high sensitivity for bioanalytical applications. In many previous studies, the affinity of nanobodies can be improved by fusing the Fc domain of human immunoglobulin G1 or connecting nanobodies to form dimerization with flexible linkers.12,13,19 Moreover, nanobodies can be produced in large quantities in prokaryotic expression systems, with a short operation cycle and are usually very stable,20 and can be atomized for direct lung delivery,19,21 which is a potential candidate for developing alternative drug delivery routes.
To further improve the affinity of nanobodies, we constructed an antibody-binding nanoplatform. Lumazine synthase isolated from the Aquifex aeolicus (AaLS) is a potential protein scaffold particle for the development of delivery and assembly vehicle (PDB:1HQK).23 AaLS is an internal hollow dodecahedron composed of 60 identical subunits, with an outer diameter of about 16 nm and an inner diameter of about 8 nm. It has the advantage of being uniform in size, symmetrical in structure, and thermally stable, with a melting temperature (Tm) value of about 120 °C.23 Due to the hollow spherical structure, AaLS is a remarkable vehicle with the surface presentation of ligands and encapsulation capability. In previous studies, modified AaLS can be used to encapsulate RNA, proteins,24,25 and also can bind small molecule drugs or antigens/antibodies to the outer surface to deliver them into cells.26 To attach nanobodies to the surface of AaLS, after genetically modifying AaLS and nanobodies by introducing SpyTag/ SpyCatcher, we conjugated the heterogeneously produced nanobodies to the preassembled AaLS scaffold by using the spontaneous isopeptide formation between SpyCatcher and its peptide partner SpyTag. Both SpyTag and SpyCatcher can be located at different positions in the protein chain and can form isopeptide bonds under various conditions (pH, buffer, and temperature).27,28 This Plug-and-Display coupling assembly strategy can avoid proteins of antibodies or antigens misfolding caused by over-long gene co-expression. The isopeptide bond-based conjugation method has also been used to design and enhance the efficacy of different particle vaccines, such as influenza A virus, HIV, cancer cells (PD-L1), etc.29–32
In summary, the study reports the development and characterization of four anti-RBD nanobodies isolated from a naïve phage display library with SARS-CoV-2 RBD. B-B2 of the four nanobodies had the most optimal neutralizing activity against pseudovirus Omicron (B.1.1.529). The attempt of attaching monovalent nanobodies to AaLS contributes to increased thermal stability and neutralizing activity of nanobodies. The results of peptide-based ELISA initially indicated that LS-B-B2 mainly reacted with some linear epitopes of the SARS-CoV-2 RBD conserved regions, LS-B-B2 could serve as a potential agent for the neutralization of SARS-CoV-2 variants.
Materials and Methods
Antigen
The antigen, SARS-CoV-2 wildtype (WT) and Omicron (B.1.1.529) spike RBD recombinant protein, were purchased from Sino Biological (Beijing. China). A DNA sequence encoding the SARS-CoV-2 WT RBD (YP_009724390.1) (Arg319-Phe541) without any terminal peptide tag. The WT RBD protein consists of 223 amino acids. A DNA sequence encoding the SARS-CoV-2 Omicron (B.1.1.529) RBD (YP_009724390.1, with mutation G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H) (Arg319-Phe541) with the Fc region of mouse IgG1 at the C-terminus. The Omicron (B.1.1.529) RBD consists of 457 amino acids. Two proteins were both expressed in HEK293 cells and dissolved in a phosphate buffer and stock at −20 °C. The experiments involving antigen, SARS-CoV-2 WT and Omicron (B.1.1.529) spike recombinant protein, only needed to be carried out under ordinary laboratory conditions.
Screening Nanobodies Against from a Naïve Phage Display Library
The phagemid vector pADL-10b was used to generate a VHH phage display library (AlpaLife) of about 2×109 cfu. E. coli SS320 cells containing the VHH library were infected with helper phage M13KO7 to produce phages displaying the encoded VHHs as fusion proteins. Phage displaying RBD (Sino Biological)-specific VHHs were biopanned as previously described.33 After 3 rounds of panning, 96 individual clones were selected and inoculated into the 2×YT medium supplemented with 100 μg/mL ampicillin at 37 °C overnight. The cell-free phage supernatant was detected by ELISA with M13Bacteriophage Antibody (HRP) (Sino Biological). P (positive OD450)/N (negative OD450) greater than 3 P/N ratios were determined as positive clones. Positive candidates were sequenced (Sangon Biotech) and aligned with CDRs amino acid sequence. A 3% bovine serum albumin (BSA) was used as a negative control for each round.
Prokaryotic Expression and Purification of Nanobodies and AaLS
An N- terminal and a C- terminal 6×His tag were added to the sequences of the nanobodies for further purification. Nanobodies are ready to be assembled with scaffolds with a 6×His tag and flexible linker (G4S)3-linked spytag003 (GenBank: MT945421.1) added at the C-terminal. The C-terminal of AaLS (PDB:1HQK) added a 6×His tag and flexible linker (GGS)4-linked spyCatcher003 (GenBank: QGX07219.1). The above sequences were all synthesized and subcloned into the pET-28a(+) vector (Generay Biotech). All the plasmids were transformed into E. coli BL21(DE3) (TransGen Biotech). Single colonies from agar plates were used to inoculate cultures of LB broth (5 mL) containing kanamycin (50 μg/mL) and grown at 37 °C overnight (250 rpm). This starter culture was then used to inoculate 200 mL LB cultures containing kanamycin (50 μg/mL) at 0.1 OD600 and grown at 37 °C with shaking (250 rpm). The expressions of all proteins were induced overnight at 18°C (200 rpm) after adding IPTG with the final concentration of 0.5 mM when the cell density reached 0.6. All proteins were purified using nickel columns (Thermo) according to the instructions. Finally, quantitating the purified proteins in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using Image J software.
Indirect ELISA to Quantitate Initial Binding
The RBD proteins were coated on a 96-well plate (Corning) at a concentration of 0.5 μg/mL, 100 mL/well at 4°C overnight, washed three times with PBST and blocked with 200 μL/well of 3% BSA blocking solution diluted with PBST for one hour at room temperature (RT). After washing three times with PBST, every well was added 100 μL of primary antibody serially diluted with blocking solution and incubated at RT for one hour. After washing three times with PBST, 100 μL/well of Anti-His tag mouse monoclonal antibody (HRP) (Atagenix) diluted 1:2000 was added and incubated for one hour at RT. After washing three times with PBST, 100 μL TMB (TIANGEN) was added to each well. After 10 minutes of color development, an equal volume of 1M HCl was added to stop the reaction, and the absorbance was read at 450nm. The 3% BSA was used as the negative control.
Forming Multivalent Nanobodies and Observed by Negative-Stain Transmission Electron Microscopy (TEM)
SpyCatcher-AaLS was reacted with 2× molar excess of nanobodies-Spytag diluted with 1M tris-buffered saline (TBS) buffer, pH 8.5, and incubated overnight at 4°C, performing SDS-PAGE to analyze the coupling efficiency. For protein shell observation, the 20 μL sample was dropped on the copper mesh grids (400 mesh), the filter paper was used to remove excess liquid from the edge of the copper mesh 3 minutes later, then stained with 3% phosphotungstic acid waited for 2–3 minutes for TEM (JEM-1400) observation. Data acquisition and processing were performed using NTA software 3.3. Every sample was recorded three times in 30s, with detection threshold 3.
Mapping Nanobody Epitopes
The ability of two nanobodies to recognize the same epitope was detected by an ELISA using RBD-coated plates. Briefly, one HRP-labeled nanobody and another nanobody without HRP were mixed, and the mixture was added to the RBD-coated plates. If the two nanobodies recognized the same epitope, they could compete to bind to the sites of RBD, thereby reducing the amount of nanobody-HRP bound to the bottom of the well and resulting in a lower OD450 value. The positive control comprised a mixture of the same labeled and unlabeled nanobodies, while the negative control was the labeled nanobody alone. Inhibition rate = (1 − OD450 nm)/ OD450 nm negative control × 100%. The inhibition rate was classified into four groups, ≥ 75% relevant; 50% to 75%, partly relevant; 25% to 50% irrelevant; < 25%, totally irrelevant. What’s more, performing molecular dynamics simulations between WT RBD and nanobody using Discovery Studio (DS). PDB files of RBD protein and four nanobodies were obtained by Phyre2.
A peptide-based ELISA was used to map the epitopes of the nanobodies. The 17 peptides were based on the protein sequence of the WT RBD, and a peptide of ZIKA EDIII (TLHGTVTVEVQYAGT) as the negative control. Each peptide ranged from 9–30 amino acids in length (GL Biochem). All peptides were provided as a lyophilized powder, reconstituted in double-distilled H2O or dimethyl sulfoxide as required to a concentration of 1 mg/mL, and stored at −20 °C. Each peptide was coated at a concentration of 1μg/mL overnight at 4 °C. The primary antibodies were diluted to the concentration of 100 μg/mL with the blocking solution. Other details are as described in indirect ELISA.
ELISA-Based SARS-CoV-2 Surrogate Virus Neutralization Test (sVNT)
To preliminarily determine the ability of nanobodies to block the binding of RBD to ACE2, ELISA-based sVNTs were performed as recommended by the manufacturer (GenScript). The serially diluted nanobodies were mixed with equal volumes of HRP-conjugated RBD and the mixture was incubated at 37°C for 30 min. 100 µL of the mixture was then added to a 96-well plate precoated with ACE2 and incubated at 37°C for 15 min. A competitive inhibitory reaction occurred between the HRP-conjugated RBD protein and the SARS-CoV-2 neutralizing antibodies. After thorough washing, the TMB substrate solution was added to the wells and incubated for 15 minutes. The reaction was stopped by adding an equal volume of stop solution and the color intensity was measured spectrophotometrically at 450 nm. The percentage of inhibition was calculated using the following formula: % inhibition = (1 − (OD450 sample/OD450 of negative control)) × 100.
Pseudovirus Neutralization Assay
All the SARS-CoV-2 pseudoviruses and the envelope recombinant plasmids are donated. The SARS-CoV-2 pseudovirus assays were performed on 293T-hACE2 cells as in the previous study.34 The 293T-hACE2 cells in pseudovirus neutralization assays were purchased from GenScript (Nanjing. China). Nanobodies were diluted serially with Dulbecco’s modified eagle medium (Gibco). 50 μL nanobodies were mixed with equal volume of 1.3×104 TCID50/mL SARS-CoV-2 WT or Omicron (B.1.1.529) pseudoviruses were incubated for 1 h in the 37 °C incubators, supplied with 5% CO2. The negative control group is composed of cells only. 100 μL freshly trypsinized 293T-hACE2 cells were added to a 96-well plate. The plate was incubated at 37 °C for 24 h. The culture supernatant was aspirated gently to leave 100 μL in each well, then, 100 μL of luciferase substrate (Beyotime) was added to each well. Two min after incubation at RT, 150 μL of lysate was transferred to white solid 96-well plates for the detection of luminescence using a multifunctional microplate detector (Synergy H1MF) to obtain the RLU to calculate IC50. All the SARS-CoV-2 pseudovirus neutralization assays were operated in the biosafety level 2 (BSL- 2) laboratory.
Surface Plasmon Resonance (SPR) Assay
Install the COOH chip following the OpenSPRTM instrument standard operating procedure. Start running at a maximum flow rate (150 µL/min) with HEPES assay buffer. After reaching the signal baseline, load 200 µL isopropyl alcohol and run for 10s to remove bubbles. After reaching the baseline, flush the sample ring with buffer and empty it with air. After the signal reaches the baseline, adjust the buffer flow to 20 µL/min. Load the EDC/NHS (1:1) solution to activate the chip. 200 µL of ligand RBD diluted in activation buffer was loaded for 4 min, and the sample loop was rinsed with HEPES and evacuated with air. Load 200 µL of Blocking Solution, rinse the sample loop with buffer HEPES and evacuated with air. Observe the baseline for 5 minutes to ensure stabilization. The analytes were diluted with buffer and loaded at 20 µL/min, and the binding time of protein and ligand was 240 s; natural dissociation was 360 s. The level of protein coupling is 5ug/mL, NaAc pH 6.0, 1150 RU. Using 10mM Glycine-HCl pH1.5 as the regeneration conditions. The analysis software used for the experimental results was TraceDrawer (Ridgeview Instruments ab, Sweden), and the analysis method was the One-to-One analysis model.
Differential Scanning Calorimetry (DSC)
The DSC measurement of samples prepared in PBS was carried out on MicroCal PEAQ-DSC (Malvern) at a heating rate of 2.00 °C per minute. To evaluate the thermal stability, the software MicroCal PEAQ-DSC software (Malvern) was used to determine the temperature with the maximum heat capacity as the apparent Tm.
Statistical Analysis
Statistical analyses were carried out using GraphPad Prism 9 (San Diego, CA, USA). All the data were presented as the mean ± SD with at least two replicates. Student’s t-test was used for two groups comparison. The Dunnett t-test was used for the analysis of the peptide‑based ELISA. Two-sided P-values < 0.05 were considered statistically significant. (*P<0.05; ****P<0.0001).
Results
Screening SARS-CoV-2 RBD Specific Nanobodies from a Naïve Alpaca VHH Library
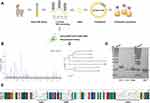
At present, there are numerous reports that single-domain antibodies have a neutralizing effect on SARS-CoV-2, which are considered potential therapeutics for COVID-19 patients.12–14 Naïve VHH library can be used for rapid acquisition and exploitation of anti-SARS-CoV-2 nanobodies through the panning using the RBD.13,35–37 On this account, we used purified RBD protein of the SARS-CoV-2 WT to perform three rounds of panning in a naïve alpaca VHH library by in vitro phage display technology (Figure 1A). After three rounds of enrichment (Table 1), 96 clones (Figure 1B) were randomly selected by phage-ELISA, and the clones with relatively high OD values were sequenced. Multiple sequence alignment and phylogenetic analyses were performed to exclude clones with identical sequences (Figure 1C).
 |
Table 1 Enrichment of Phage Particles Against SARS-CoV-2 RBD-Specific Nanobodies During Three Rounds of Panning |
Four of the nanobodies with different amino acid sequences of CDR3 were expressed successfully by the prokaryotic expression system with E. coli strain BL21(DE3). SDS-PAGE showed the recombinant proteins were detected at the expected apparent molecular weights, which was 15~17 kDa (Figure 1D and E).
Affinity and Neutralizing Potency of Monovalent Nanobodies
Due to the potency of nanobodies being greatly affected by their affinity to antigen,15,38 The binding kinetics of the four nanobodies were measured using SPR assays with the purified WT and Omicron (B.1.1.529) RBD, respectively. The results showed that the four nanobodies have affinities to WT RBD with KD values ranging from 3.37 nM to 317 nM. Except for B2-3, the affinities of the other three nanobodies to Omicron (B.1.1.529) RBD maintained, while B-B2 has the highest affinity to Omicron (B.1.1.529) RBD, KD value is 49 nM (Table 2).
 |
Table 2 The Results of Binding Affinity Between Nanobodies and RBD Using SPR |
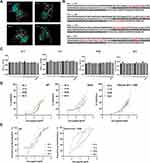
To characterize the binding epitopes between the four nanobodies and RBD, we performed molecular docking between nanobodies and RBD using the computer molecular simulation software DS to predict and analyze the antigen-binding epitopes of nanobodies (Figure 2A). It can be seen from the red part of the RBD sequence in the prediction model that there is a binding epitope of up to 13 or 11 amino acids between B1-4 or B-B2 and RBD (Figure 2B). Except for B2-3, the predicted binding epitopes of these nanobodies are not cover the RBM region (marked in gray). As the results shown in the competitive assays between nanobodies (Table 3), B-B2 and C1-5 were relevant, for which the inhibition rate was 77.22%. B-B2 and B1-4, B-B2 and B2-3 were partly relevant, with inhibition rates of 55.86% and 61.78%, respectively. The inhibition rate between the remaining combinations is less than 50%. We also tried to identify the binding epitopes of nanobodies by peptide‑based ELISA, however, no statistically significant binding activities were observed between nanobodies and synthetic peptides (Figure 2C).
 |
Table 3 Inhibition Rate Between Two Nanobodies |
To assess the neutralizing activity of these nanobodies, we carried out the sVNTs that competed with hACE2 for RBD and the pseudovirus neutralization assays. The neutralizing activity of nanobodies against authentic SARS-COV-2 in vivo was not performed in the present study, because SARS-COV-2 live virus neutralization assays must be carried out in a biosafety level-3 laboratory and are time-consuming. The ELISA-based sVNT could be used to assess the ability of the nanobodies to block RBD-ACE2 interaction, and is more efficient and safer, without using viruses, and the detection can be completed in a short time under common experimental conditions. Moreover, the result of sVNTs is closely related to the cell-based neutralization assays.39,40 The pseudovirus neutralization assay based on the cell entry pattern similar to that of authentic viruses is also an important supplementary assay of antiviral activity.
The results of sVNTs showed that all four nanobodies could block WT RBD binding to ACE2 in a dose-dependent manner (Figure 2D). Then, we carried out the sVNTs using the Delta and Omicron (B.1.1.529) RBD, and found that the %inhibition could be maintained well (Figure 2D). We also carried out the SARS-CoV-2 WT and Omicron (B.1.1.529) pseudovirus neutralization assays with the nanobodies. The pseudovirus luciferase assay results indicated that four nanobodies neutralized WT SARS-CoV-2 pseudotyped virus with an IC50 value at approximately 4.889 µg/mL to 11.49 µg/mL. Furthermore, although the previously dominant Omicron variant holds 15 unprecedented mutations in its RBD, the nanobodies neutralized Omicron (B.1.1.529) pseudotyped virus with an IC50 value at approximately 1.658 µg/mL to 16.58 µg/mL. B-B2 showed the highest activity with an IC50 at 1.658 µg/mL to neutralize Omicron (B.1.1.529) pseudotyped viruses (Figure 2E and Table 4).
 |
Table 4 The IC50 of Monovalent and Multivalent Nanobodies in the Pseudovirus Neutralization Assays |
Development and Characterization of Multivalent Nanobodies with Thermostable Scaffold AaLS
Previously, it is reported that the binding affinity of the monomer nanobody can be improved by tandem linking.13,35 To further improve the affinity and neutralizing activity of nanobodies, we tried to develop multivalent nanoantibodies with SpyCatcher-modified AaLS as shown in Figure 3A. AaLS were expressed in E. coli in soluble form and purified. To confirm that the AaLS can maintain its ability to self-assemble into dodecahedral nanostructures, we observed the structural characteristics of the SpyCatcher-conjugated nanoparticles by negative staining TEM. As shown in Figure 3C, the SpyCatcher-modified AaLS can still form a dodecahedral sphere.
Next, we tried to assemble the scaffolds and nanobodies at different molar ratios of 1:2, and 1:1 in TBS buffer. Taking the nanobody B1-4 as an example, the SDS-PAGE showed (Figure 3B) that when nanobodies were excessive, the number of remaining scaffolds decreases obviously and more scaffolds were bound. To allow the subunits of the AaLS to be decorated with nanobodies as much as possible, we used the reaction condition of a 2:1 molar ratio and then filtered out the nanobodies that were not bound to Alas by the ultrafiltration tube. The assembly products were observed by negative-stain TEM. As we expected, the diameter of the assembled product was larger than that of the AaLS without nanobodies, and the spherical structure was maintained, which indicated that nanobodies were successfully attached to the scaffolds (Figure 3C).
The Multivalent Nanobodies are Highly Potent
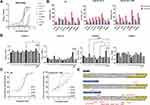
We then characterize the affinity of the assembled products of scaffolds and nanobodies by using an indirect ELISA. Taking B1-4 as an example, we named the assembled products LS-B1-4. The indirect ELISA results indicated that the binding affinity of the multivalent nanobodies was significantly higher than that of monomer nanobodies (Figure 4A).
To compare neutralizing activity between monomer and multivalent nanobodies, sVNTs were carried out. The results showed that the %inhibition of multivalent nanobodies was higher than that of monomer nanobodies at the same concentration (Figure 4B). In the pseudovirus neutralization assays, all the multivalent nanobodies exhibited better neutralizing ability compared to the monovalent form. Results showed that LS-B-B2 exhibited the highest neutralizing ability with an IC50 at 0.653 μg/mL against pseudotyped SARS-CoV-2 Omicron (B.1.1.529) (Figure 4C), was improved compared to the monovalent form B-B2 (IC50 = 1.658 μg/mL) (Figure 2E).
The peptide‑based ELISA was used to explore the epitopes of four multivalent nanobodies binding to WT RBD (Figure 4D). A total of 17 WT RBD peptides (Table 5) were selected and a Zika EDIII peptide was used as a negative control. It was found that compared with the control peptide, LS-B1-4 mainly recognized peptides 10 and 16, LS-C1-5 mainly recognized peptide 16, and LS-B-B2 recognized peptides 5, 8, 12, and 16. Of note is that peptides 5 (S381-403 GVSPTKLNDLCFTNVYADSFVIR) and 16 (S519-543 HAPATVCGPKKSTNLVKNKCVNFNF) are conserved across the variants of SARS-COV-2 Delta and Omicron (B.1.1.529) (Figure 4E), which may initial clarify the mechanisms of these three nanobodies maintaining the neutralizing potency of SARS-COV-2 variants.
 |
Table 5 17 Peptides of SARS-COV-2 WT RBD |
Improved Thermal Stability of Multivalent Nanobodies
While nanobody is often heat resistant with the Tm from 50 to 80°C, it is not heatproof,41–43 obstructing it as a superior reagent for detection, diagnostic, and therapeutic applications. Many nanobodies have been engineered to increase their Tm, making them stable until exposed to extreme temperatures. Since AaLS isolated from the hyper-thermophile has excellent robustness, the scaffold is expected to improve the thermal stability of monovalent nanobodies. Taking B2-3 as an example, DSC analysis showed that the Tm of B2-3 was about 50°C, and that of LS-B2-3 was about 70°C, being increased by 20 degrees (Figure 5A).
While the DSC analysis could directly measure the temperature at which the nanobodies lose their secondary structure, it is necessary to measure the activity after exposure to high temperature, since the change of binding affinity is not parallel to the result of physical parameters in some cases.20 Therefore, an indirect ELISA and SDS-PAGE were used to observe the binding affinity of nanobodies after one hour of treatment at different temperature conditions (no heat treatment, 50°C, 70°C, 90°C) for an hour (Figure 5B and C). The results of indirect ELISA showed that the binding ability of B2-3 degraded obviously with the increase of temperature (no heat treatment to 70°C), while the affinity of 90°C treated B2-3 was higher than that of B2-3 without heat treatment. Although the B2-3 at 90°C has been degraded partially as observed by SDS-PAGE. By contrast, 90°C did not significantly compromise the affinity of the multivalent nanobody, LS-B2-3, it was more thermally stable and retained good binding affinity, and the protein observed by SDS-PAGE did not been substantially reduced.
In summary, our research manifests that the nanobodies attached to scaffolds are endowed with higher Tm, greater ability to refold after denaturation, and ability to function after heat exposure.
Discussion
COVID-19 has brought unprecedented threats to human health and become a global concern. The variants of SARS-CoV-2 compromise the efficacy of most vaccines or therapeutic monoclonal antibodies. High-throughput naïve library can be used as an available platform for quickly obtaining antibodies against specific antigens. To date, some nanobodies screened from naïve nanobody libraries have neutralization potencies for SARS-CoV-2.35,44 Here, we developed four nanobodies specific to the SARS-CoV-2 RBD and used the prokaryotic expression system to express them successfully. Since the nanobodies were isolated from nonimmune synthetic libraries, the binding affinity and neutralizing activity needed to be further improved. We modified the nanobodies.
Protein cage nanoparticles isolated from AaLS have highly uniform nano-size and symmetrical structure, and they have genetic and chemical plasticity amenable to simultaneously introducing multiple cell-specific targeting ligands at desired sites according to the purpose. At present, they have been widely explored and utilized as effective delivery nanoplatforms for diagnosis and treatment in the biomedical field. AaLS is the expected antibody-mediated targeted delivery nanoplatform for us to modify antibodies to increase affinity. Since the spike proteins naturally exist as a trimer on the surface of SARS-CoV-2, there is the possibility that the multimerized nanobodies could target more than one RBD antigen at the same time, and the multimeric nanobody-antigen structure could be more stable than the monomeric nanobody-antigen complex. Consistent with the assumption, in the present study, we observed the LS-nanobodies showed better potency than monoclonal antibodies in indirect ELISA, the sVNTs, and the pseudovirus neutralizing assays.
In addition, since the nanoplatform is isolated from hyperthermophile Aquifex aeolicus, it may contribute to the thermal stability of the nanobodies. In the study, we observed that the nanobodies attached to AaLS can maintain binding affinity after being treated at different temperatures. The affinity of monovalent nanobodies for antigen decreased with the increase of treatment temperature. However, the affinity of monovalent nanobody B2-3 treated after 90°C is a special case. It is possible that the spatial structure of B2-3 was reconstructed due to heat-induced denaturation that exceeded its melting point, which transformed the conformational space of a CDR, opened the hidden side of its binding to the antigen, and increased the binding affinity.22 In a word, the AaLS could be used as a promising candidate for multifunctional delivery nanoplatform. Its advantages will further increase its application in disease diagnosis, prevention, and treatment.
The ACE2 receptor-binding motif (RBM) contains the majority of RBD mutations found in Omicron (B.1.1.529). The RBM binders, usually competing with ACE2 to bind to RBD, lose (or weaken) their binding and neutralizing activities to the SARS-CoV-2 variants, especially the most recent Omicron.45 Previous studies have revealed the presence of conserved epitopes located on SARS-CoV-2 RBDs and the Omicron neutralizing potency of non-RBM antibodies.46,47 In this study, we explored the conservation of the nanobodies epitope on the RBD using DS molecular simulation docking. The results showed that the binding sites of B-B2 do not cover the RBM region. The peptide‑based ELISA indicated that it mainly recognized the peptides 5 (S381-403) and 16 (S519-543), and these two peptides were 100% conserved across the variants of SARS-COV-2, suggesting the nanobodies recognize them might not lose most of the affinity. These results preliminarily indicated the region of antigen recognized by B-B2 were conserved so that B-B2 can maintain the neutralizing activity against Omicron (B.1.1.529). This nanobody combined with conserved epitopes, might also be a powerful agent for creating effective broad-spectrum antiviral agents. Besides, though the KD of B2-3 was the highest in SPR using WT RBD, it did not show a better neutralizing activity. However, the neutralizing activity is not only simply determined by the affinity of the antibody for antigen binding, it is also affected by a variety of factors, such as the heterogeneity of the antibody, the antigenic determinant of the antibody targeting, and the property of binding to complement.48
There are some limitations in this research. Firstly, the SARS-COV-2 live virus neutralization assay is the gold standard for measuring antibody neutralization ability. However, it must be carried out in a biosafety level-3 laboratory. Therefore, here, we performed the pseudovirus neutralization assay, which is also suitable for simulating the inhibition of nanobodies that inhibits the virus entry. Secondly, the mechanism of these three nanobodies maintaining pseudovirus neutralizing activity to variant strains deserves further investigation. However, the pattern of antigen-antibody interaction is very complex. In our study, the binding site prediction of the nanobody-RBD complex relies on peptide-ELISA and DS simulation docking, which lacked cryo-electron microscopy or X-ray diffraction to investigate the crystal structure parameters.
Conclusion
In summary, our study developed four nanobodies against SARS-COV-2 WT RBD from a non-immunized alpaca VHH library which may target epitopes of conserved regions of antigens, and improved the nanobodies using AaLS. Of them, LS-B-B2 exhibited the highest neutralizing ability against pseudotyped SARS-CoV-2 Omicron (B.1.1.529). Our study indicated that the AaLS could be used as an antibody-binding nanoplatform to present nanobodies on its surface and improve the potency of nanobodies. The multivalent nanobody LS-B-B2 may serve as a potential agent for the neutralization of SARS-CoV-2 variants.
Ethics Approval
Ethics committee approval was not required as IRB approval is not a requisite for this study.
Acknowledgments
We thank AlpaLife for their excellent technical assistance. We also thank Bixia Hong and Pro. Huahao Fan (Beijing University of Chemical Technology) for the kind gift of the SARS-CoV-2 pseudoviruses and plasmids. This work was funded by the National Key Research and Development Project (2018YFE0208000), the Shenzhen Science and Technology Program (JSGG20220606142207017), the Guangdong Province Drug Administration Science and Technology Innovation Project (2022ZDZ12), the Central Government Guides Local Science and Technology Development Funds to Freely Explore Basic Research Project (2021Szvup171), the State Key Program of National Natural Science of China (U1808202), NSFC International (regional) cooperation and exchange program (31961143024), and the Key Program of Inner Mongolia (2019ZD006).
Disclosure
Dr Yuying Lu reports a patent ZLTA20220905 pending. The authors report no other conflicts of interest in this work.
References
1. Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi:10.1038/s41586-021-03398-2
2. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi:10.1038/s41586-021-03324-6
3. Planas D, Saunders N, Maes P, et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602(7898):671–675. doi:10.1038/s41586-021-04389-z
4. Dejnirattisai W, Huo J, Zhou D, et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467–484.e415. doi:10.1016/j.cell.2021.12.046
5. Wang P, Casner RG, Nair MS, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747–751.e744. doi:10.1016/j.chom.2021.04.007
6. McCallum M, Walls AC, Sprouse KR, et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374(6575):1621–1626. doi:10.1126/science.abl8506
7. Yuan M, Huang D, Lee CD, et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373(6556):818–823. doi:10.1126/science.abh1139
8. Dejnirattisai W, Zhou D, Ginn HM, et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184(8):2183–2200.e2122. doi:10.1016/j.cell.2021.02.032
9. McCallum M, Czudnochowski N, Rosen LE, et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science. 2022;375(6583):864–868. doi:10.1126/science.abn8652
10. Hastie KM, Li H, Bedinger D, et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374(6566):472–478. doi:10.1126/science.abh2315
11. Wang N, Sun Y, Feng R, et al. Structure-based development of human antibody cocktails against SARS-CoV-2. Cell Res. 2021;31(1):101–103. doi:10.1038/s41422-020-00446-w
12. Schoof M, Faust B, Saunders RA, et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370(6523):1473–1479. doi:10.1126/science.abe3255
13. Huo J, Le Bas A, Ruza RR, et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol. 2020;27(9):846–854. doi:10.1038/s41594-020-0469-6
14. Koenig PA, Das H, Liu H, et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371:6530. doi:10.1126/science.abe6230
15. Wrapp D, De Vlieger D, Corbett KS, et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181(5):1004–1015.e1015. doi:10.1016/j.cell.2020.04.031
16. Jovčevska I, Muyldermans S. The Therapeutic Potential of Nanobodies. BioDrugs. 2020;34(1):11–26. doi:10.1007/s40259-019-00392-z
17. Hu Y, Liu C, Muyldermans S. Nanobody-based delivery systems for diagnosis and targeted tumor therapy. Front Immunol. 2017;8:1442. doi:10.3389/fimmu.2017.01442
18. Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi:10.1146/annurev-biochem-063011-092449
19. Xiang Y, Nambulli S, Xiao Z, et al. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370(6523):1479–1484. doi:10.1126/science.abe4747
20. Dumoulin M, Conrath K, Van Meirhaeghe A, et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11(3):500–515. doi:10.1110/ps.34602
21. Esparza TJ, Martin NP, Anderson GP, Goldman ER, Brody DL. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci Rep. 2020;10(1):22370. doi:10.1038/s41598-020-79036-0
22. Ikeuchi E, Kuroda D, Nakakido M, Murakami A, Tsumoto K. Delicate balance among thermal stability, binding affinity, and conformational space explored by single-domain V(H)H antibodies. Sci Rep. 2021;11(1):20624. doi:10.1038/s41598-021-98977-8
23. Zhang X, Meining W, Fischer M, Bacher A, Ladenstein R. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons. J Mol Biol. 2001;306(5):1099–1114. doi:10.1006/jmbi.2000.4435
24. Wörsdörfer B, Pianowski Z, Hilvert D. Efficient in vitro encapsulation of protein cargo by an engineered protein container. J Am Chem Soc. 2012;134(2):909–911. doi:10.1021/ja211011k
25. Seebeck FP, Woycechowsky KJ, Zhuang W, Rabe JP, Hilvert D. A simple tagging system for protein encapsulation. J Am Chem Soc. 2006;128(14):4516–4517. doi:10.1021/ja058363s
26. Ra JS, Shin HH, Kang S, Do Y. Lumazine synthase protein cage nanoparticles as antigen delivery nanoplatforms for dendritic cell-based vaccine development. Clin Exp Vaccine Res. 2014;3(2):227–234. doi:10.7774/cevr.2014.3.2.227
27. Zakeri B, Fierer JO, Celik E, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 2012;109(12):E690–697. doi:10.1073/pnas.1115485109
28. Li L, Fierer JO, Rapoport TA, Howarth M. Structural analysis and optimization of the covalent association between SpyCatcher and a peptide Tag. J Mol Biol. 2014;426(2):309–317. doi:10.1016/j.jmb.2013.10.021
29. Tan TK, Rijal P, Rahikainen R, et al. A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat Commun. 2021;12(1):542. doi:10.1038/s41467-020-20654-7
30. Rahikainen R, Rijal P, Tan TK, et al. Overcoming symmetry mismatch in vaccine nanoassembly through spontaneous amidation. Angew Chem Int Ed Engl. 2021;60(1):321–330. doi:10.1002/anie.202009663
31. Bruun TUJ, Andersson AC, Draper SJ, Howarth M. Engineering a rugged nanoscaffold to enhance plug-and-display vaccination. ACS Nano. 2018;12(9):8855–8866. doi:10.1021/acsnano.8b02805
32. Brune KD, Leneghan DB, Brian IJ, et al. Plug-and-display: decoration of virus-like particles via isopeptide bonds for modular immunization. Sci Rep. 2016;6:19234. doi:10.1038/srep19234
33. Li Q, Zhang F, Lu Y, et al. Highly potent multivalent VHH antibodies against Chikungunya isolated from an alpaca naïve phage display library. J Nanobiotechnology. 2022;20(1):231. doi:10.1186/s12951-022-01417-6
34. Nie J, Li Q, Wu J, et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat Protoc. 2020;15(11):3699–3715. doi:10.1038/s41596-020-0394-5
35. Lu Q, Zhang Z, Li H, et al. Development of multivalent nanobodies blocking SARS-CoV-2 infection by targeting RBD of spike protein. J Nanobiotechnology. 2021;19(1):33. doi:10.1186/s12951-021-00768-w
36. Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi:10.1038/s41586-020-2381-y
37. Hanke L, Vidakovics Perez L, Sheward DJ, et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat Commun. 2020;11(1):4420. doi:10.1038/s41467-020-18174-5
38. Li W, Schäfer A, Kulkarni SS, et al. High potency of a bivalent human V(H) Domain in SARS-CoV-2 animal models. Cell. 2020;183(2):429–441.e416. doi:10.1016/j.cell.2020.09.007
39. Tan CW, Chia WN, Qin X, et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38(9):1073–1078. doi:10.1038/s41587-020-0631-z
40. Abe KT, Li Z, Samson R, et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5:19. doi:10.1172/jci.insight.142362
41. Graef RR, Anderson GP, Doyle KA, et al. Isolation of a highly thermal stable lama single domain antibody specific for Staphylococcus aureus enterotoxin B. BMC Biotechnol. 2011;11:86. doi:10.1186/1472-6750-11-86
42. Turner KB, Zabetakis D, Legler P, Goldman ER, Anderson GP. Isolation and epitope mapping of staphylococcal enterotoxin B single-domain antibodies. Sensors. 2014;14(6):10846–10863. doi:10.3390/s140610846
43. Olson MA, Legler PM, Zabetakis D, Turner KB, Anderson GP, Goldman ER. Sequence tolerance of a single-domain antibody with a high thermal stability: comparison of computational and experimental fitness profiles. ACS Omega. 2019;4(6):10444–10454. doi:10.1021/acsomega.9b00730
44. Li Q, Humphries F, Girardin RC, et al. Mucosal nanobody IgA as inhalable and affordable prophylactic and therapeutic treatment against SARS-CoV-2 and emerging variants. Front Immunol. 2022;13:995412. doi:10.3389/fimmu.2022.995412
45. VanBlargan LA, Errico JM, Halfmann PJ, et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022;28(3):490–495. doi:10.1038/s41591-021-01678-y
46. Li C, Zhan W, Yang Z, et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell. 2022;185(8):1389–1401.e1318. doi:10.1016/j.cell.2022.03.009
47. Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi:10.1126/science.abb7269
48. Weidenbacher PA, Waltari E. Converting non-neutralizing SARS-CoV-2 antibodies into broad-spectrum inhibitors. Nat Chem Biol. 2022;18(11):1270–1276. doi:10.1038/s41589-022-01140-1
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.