Back to Journals » International Journal of Nanomedicine » Volume 14
A genipin-crosslinked protein–polymer hybrid system for the intracellular delivery of ribonuclease A
Authors Liang X, Tang X, Yang J, Zhang J, Han H , Li Q
Received 29 March 2019
Accepted for publication 27 July 2019
Published 10 September 2019 Volume 2019:14 Pages 7389—7398
DOI https://doi.org/10.2147/IJN.S210486
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Xiao Liang, Xiuhui Tang, Jiebing Yang, Jiayuan Zhang, Haobo Han, Quanshun Li
Key Laboratory for Molecular Enzymology and Engineering of Ministry of Education, School of Life Sciences, Jilin University, Changchun 130012, People’s Republic of China
Correspondence: Haobo Han; Quanshun Li
Key Laboratory for Molecular Enzymology and Engineering of Ministry of Education, School of Life Sciences, Jilin University, Changchun 130012, People’s Republic of China
Tel +86 4 318 515 5201
Fax +86 4 318 515 5200
Email [email protected]; [email protected]
Background: Therapeutic proteins have been widely used in the treatment of various diseases, and effective carriers are highly required for achieving protein delivery to obtain favorable treatment potency.
Materials and methods: A protein–polymer hybrid system was constructed through the genipin-mediated crosslinking of polyethyleneimine with a weight-average molecular weight of 25,000 g/mol (PEI25K) and ribonuclease A (RNase A), namely RGP.
Results: The RGP nanoparticles were observed to be easily internationalized in HeLa cells owing to the introduction of positively charged PEI25K, thereby triggering the antiproliferative effects by cleaving RNA molecules in the tumor cells. Moreover, red fluorescence could be obviously visualized in the tumor cells after RGP delivery, which was attributed to the intrinsic characteristics of genipin.
Conclusion: The protein–polymer hybrid system prepared via the genipin-mediated crosslinking has exhibited potential to be used as a theranostic platform for both in vivo imaging and delivering diverse therapeutic proteins.
Keywords: genipin, polyethyleneimine, ribonuclease A, protein delivery, antitumor efficacy
Introduction
In the past two decades, a series of therapeutic proteins or peptides including growth factors, cytokines, antibodies and enzymes have been successfully developed due to the rapid progress of biotechnological techniques.1,2 Meanwhile, protein-based therapy has exhibited great potential in the treatment of various diseases owing to the characteristics of high pharmacological potency and low toxicity.3,4 Among them, the cytotoxic ribonuclease A (RNase A) could achieve the cleavage of the intracellular RNA molecules and induce the cell apoptosis, which has been demonstrated to possess favorable killing ability against tumor cells.5–10 Nevertheless, it is still a great challenge to achieve an effective bioavailability and clinic applications of proteins, mainly attributed to their low stability, easy protease degradation and poor membrane permeability.11–13
Encouraged by the recent development of nanotechnology, the nanocarriers including inorganic nanoparticles, cationic lipids, protamine, peptides and polymers have provided indispensable tools for the intracellular delivery of proteins, yielding an improvement of stability, permeability and bioavailability of cargoes.11,13–22 Particularly, polyethyleneimine (PEI) has been widely employed as gene carriers as its amino-rich structure could provide a high density of positive charge and further promote the cellular uptake through the electrostatic interaction with the negatively charged cell membrane.23,24 In addition, it could facilitate the lysosomal escape through “proton sponge” effect and protect the payload from the degradation in the acidic and enzymatic environment of endo/lysosomes.25,26 In our previous report, PEI25K was successfully crosslinked with thermophilic histone through genipin to prepare a protein–polymer hybrid gene carrier, which showed favorable biocompatibility and excellent transfection efficiency owing to the synergistic effects between these two components.27 In this system, genipin, which is the enzymatic product of geniposide from the fruit of gardenia plane, was used as a crosslinking agent possessing favorable activity with primary amine groups and colorimetric and fluorogenic activity.28–30 Thus, we infer that the genipin-mediated crosslinking will be a powerful tool to construct protein–polymer hybrid systems for realizing the delivery of therapeutic proteins.
Herein, a protein–polymer hybrid system was synthesized through the genipin-mediated crosslinking of PEI25K and RNase A for realizing the intracellular delivery of RNase A, namely RGP (Scheme 1). As RNase A is a therapeutic protein with favorable antitumor efficacy,5–10 the intracellular delivery and the further antiproliferative effects of RGP were systematically evaluated.
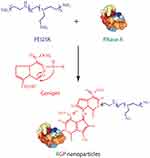 |
Scheme 1 The synthetic approach of RNase A-PEI25K hybrid system (RGP) via genipin-mediated crosslinking. |
Materials and methods
Materials
Branched PEI25K (impurities: ≤1% water) and bovine pancreatic RNase A (≥70 kU/mg protein) were purchased from Sigma-Aldrich (St. Louis, MS, USA). Genipin (>98%) was provided by Zhixin Biotechnol. Co. (Linchuan, China). RNaseAlert® kit was obtained from Integrated DNA Technologies, Inc. (Coralville, IA, USA). FBS and DMEM were purchased from Kangyuan Co. (Beijing, China) and Gibco (Grand Island, NE, USA), respectively. BCA protein assay kit was provided by BioTeke Co. (Beijing, China). Blue plus II protein marker was purchased from TransGen Biotech. (Beijing, China). BSA and MTT were purchased from Amresco (Solon, OH, USA). LIVE/DEAD® Viability/Cytotoxicity kit and one-step TUNEL cell apoptosis detection kit were obtained by Thermo Fisher (Grand Island, NE, USA) and Beyotime (Jiangsu, China), respectively. The Annexin V-FITC/PI apoptosis detection kit was provided by Vazyme Co. (Nanjing, China).
Preparation and characterization of RGP
Briefly, RNase A (0.01 μmol) was mixed with 5 mL of PEI25K solution (0.10 μmol), and genipin (0.01 μmol) was added into the mixture. After stirring at 4°C for 24 hrs, the samples were dialyzed against distilled water for 24 hrs to remove the excess genipin (MWCO: 3500 Da). The product RGP was obtained through lyophilization and then subjected to systematic characterization. The Fourier-transformed infrared spectrometry (FTIR) spectra were recorded in the range of 4000–600 cm−1 using KBr pellets on a Bruker V70 FTIR spectrometer. The UV-Vis spectra were conducted on a Shimadzu 2700 spectrophotometer in the wavelength range of 190–350 nm. The MALDI-TOF mass spectra were conducted on an AB SCIEX 5800 mass spectrometer. SDS-PAGE was conducted on 15% polyacrylamide gel with 15 μg protein per well (80 V, 150 mins), in which the concentration of RNase A in RGP was measured using BCA protein assay kit. Far UV circular dichroism spectra were performed on a JASCO 810 instrument in the range of 190–250 nm with a scanning speed of 100 nm/min, in which 1 mg/mL of RNase A concentration in distilled water was used for analysis. The transmission electron microscopy (TEM) image of RGP nanoparticles was captured on a HITACHI-H800 microscope at an accelerating voltage of 200 kV. The hydrodynamic diameter and zeta potential of RGP nanoparticles were determined using a Malvern Nano ZS90 Zetasizer (Malvern, UK).
Enzymatic activity assay
The enzymatic activities of RNase A and RGP were measured using RNaseAlert® kit according to the manufacturer’s protocol. 10 μL of RNaseAlert® substrate was first mixed with 10 μL of assay buffer provided in the kit. Then, 80 μL of RNase A or RGP solution with RNase A concentration of 1 μg/mL was added into the above solution individually. The fluorescence intensity at 520 nm (excited at 490 nm) was monitored within 30 mins.
Intracellular uptake analysis
The HeLa cells were obtained from the Shanghai Institute of Cell Bank (Shanghai, China) and inoculated in 6-well plates at a density of 3.0×105 cells/well. After the culture at 37°C overnight prior to the transfection, the medium was removed, and the cells were incubated with RGP at different concentration (0.5–8.0 μg/mL RNase A) in FBS-free DMEM for 4 hrs. Then, the medium was discarded, and the cells were collected, suspended in 1 mL PBS and analyzed through Calibur flow cytometry (BD Bioscience, Mountain View, USA) with excitation and emission wavelengths of 488 and 575 nm, respectively. For the confocal laser scanning microscope (CLSM) analysis, HeLa cells were seeded into 6-well plates with sterilized coverslips at a density of 2.2×105 cells/well and cultured at 37°C overnight. Subsequently, 2 mL of FBS-free DMEM was added into each well after removing the medium, and the cells were transfected with 4 μg/mL of RGP solution for 6 hrs. The medium was discarded after the transfection, and the cells were washed with PBS three times, fixed with 4% paraformaldehyde solution for 15 mins and stained with DAPI solution (1 μg/mL) for 5 mins. Finally, the coverslips were observed on an LSM 710 CLSM (Carl Zeiss Microscopy LLC, Jena, Germany).
Endosomal escape analysis of RGP nanoparticles
The HeLa cells were inoculated in a 6-well plate with a sterilized coverslip at an initial density of 2.5×105 cells/well and cultured at 37°C overnight. Subsequently, the cells were incubated with 1 mL FBS-free DMEM containing RGP nanoparticles (RNase A concentration of 4 μg/mL) for 2 and 6 hrs, respectively. Afterward, the cells were stained with LysoTracker Green DND-26 (ThermoFisher, Eugene, OR) for 5 mins as described in previous reports.31–33 After washing with PBS three times, the cells were fixed with 4% paraformaldehyde solution for 15 mins and stained with DAPI solution (1 μg/mL) for 5 mins. Finally, the coverslip was subjected to the analysis on an LSM 710 CLSM (Carl Zeiss Microscopy LLC to detect the endosomal escape of RGP nanoparticles.
In vitro antiproliferative effect assay
The MTT assay was employed to evaluate the in vitro antiproliferative effect of RNase A and RGP, using 0.1% Triton X-100 as the positive control according to previous reports.18,34 Briefly, 200 μL of HeLa cell suspension was added into 96-well plates at a density of 8,000 cells/well and incubated at 37°C overnight. The cells were then incubated with 0.1% Triton X-100, PEI25K, RNase A and RGP with different RNase A concentrations. After the treatment for 24 hrs, 20 μL of MTT solution (5 mg/mL) was added into each well, and the cells were incubated for an additional 4 hrs. Subsequently, the MTT solution was removed and dimethyl sulfoxide (200 μL) was added into each well to dissolve the formazan crystals. The absorbance at 570 nm was measured using HBS-1906A microplate reader (Nanjing, China), and the cell viability was calculated based on the absorbance values of treated and untreated groups.
Live/dead staining
Briefly, HeLa cells were seeded in 6-well plates at a density of 2.2×105 cells/well and incubated at 37°C overnight. The cells were treated with 0.1% Triton X-100, PEI25K, RNase A and RGP for 48 hrs (RNase A concentration of 4 μg/mL) and washed with PBS twice. According to the manufacturer’s instructions, the cells were stained with live/dead staining reagents for 30 mins, washed with PBS three times and observed on an IX71 fluorescence microscopy (Olympus, Tokyo, Japan).
Cell apoptosis analysis
The HeLa cells were seeded into 6-well plates at a density of 2.2×105 cells/well and cultured at 37°C overnight. Then, the cells were treated with 0.1% Triton X-100, PEI25K, RNase A and RGP for 48 hrs (RNase A concentration of 4 μg/mL). According to the manufacturer’s protocol, the collected cells were treated with the corresponding solutions in the kit, and the cell apoptosis was assayed on a Calibur flow cytometry (BD Bioscience).
TUNEL assay
The HeLa cells were seeded into 6-well plates at a density of 2.2×105 cells/well and cultured at 37°C overnight. After treating with 0.1% Triton X-100, PEI25K, RNase A and RGP for 48 hrs (RNase A concentration of 4 μg/mL), the cells were washed with PBS twice and fixed with 75% ethanol for 30 mins. Followed by rinsing with PBS three times, the cells were suspended in PBS containing 0.3% Triton X-100 and incubated at room temperature for 5 mins. According to the manufacturer’s protocol, the cells were incubated with TUNEL detection solution in the kit for 1 hr, washed with PBS three times and detected using an IX71 fluorescence microscopy (Olympus).
Statistical analysis
Data were presented as mean value ± SD, and statistical significance of differences was analyzed by one-way ANOVA using SPSS Statistics 23.0 complemented with Student’s t-test (n.s., not significant; *p<0.05; **p<0.01).
Results and discussion
The protein–polymer hybrid system RGP was synthesized through the genipin-mediated crosslinking of RNase A and PEI25K (Scheme 1), an aglycon of geniposide which has been widely used as a natural crosslinking agent.27,30 Then, the structure of RGP was characterized by FTIR, as shown in Figure 1. Apparently, the bands of RGP generated from RNase A were observed for protein amide I band at 1639 cm−1 and N-H stretching vibration at 3338 cm−1, while the characteristic bands from the component PEI25K were confirmed to be C-H stretching vibration at 2938 and 2810 cm−1, C-H bending vibration at 1455 cm−1, N-H stretching vibration at 3277 cm−1 and C-N stretching vibration at 1115 and 1046 cm−1. Therefore, the FTIR spectra clearly demonstrated the presence of characteristic peaks of both RNase A and PEI25K in the product RGP. Moreover, compared with RNase A, the maximum absorption wavelength of RGP in the UV-Vis spectrum could be observed to be slightly red shifting (Figure 2). As there was no absorbance in the UV-Vis spectra of PEI25K, genipin and the product from genipin-mediated crosslinking of PEI25K (Figure S1), the red shift of RGP was probably caused by the structural change of RNase A induced by the genipin crosslinking. Further, the molecular weight of RGP was determined by MALDI-TOF mass spectrum and SDS-PAGE analysis. As shown in Figure 3, the molecular weight of RNase A was 13.7 kDa, and the value of RGP increased to 66.3 kDa, indicating that RNase A was successfully crosslinked with PEI25K. Similar results could be observed in SDS-PAGE analysis, in which a major band with a molecular weight of 66.0 kDa was detected for the sample RGP (Figure 4). Interestingly, different from the single band of RNase A, lagging bands with different molecular weights could be found for RGP. This phenomenon was probably caused by the genipin-mediated crosslinking of RNase A and PEI25K in various molar ratios. In addition, a mixture of chains with different molecular weights in PEI25K could also contribute to these results. Overall, all these results clearly confirmed the successful crosslinking of PEI25K and RNase A mediated by genipin.
 |
Figure 1 FTIR spectra analysis of PEI25K, RNase A and RGP. |
 |
Figure 2 UV-Vis spectra of RNase A and RGP. |
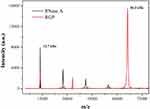 |
Figure 3 The MALDI-TOF mass spectra of RNase A and RGP. |
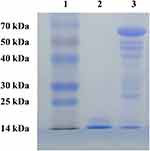 |
Figure 4 SDS-PAGE analysis of RNase A and RGP. Lane 1: marker; lane 2: RNase A; and lane 3: RGP. |
Subsequently, the morphology, hydrodynamic diameter and zeta potential of RGP were characterized. As shown in Figure S2, TEM image presented the spherical structure of RGP nanoparticles, with a particle size of ca. 200 nm. Then, the hydrodynamic diameter and zeta potential values of RGP nanoparticles were determined to be 193.1±18.3 nm and +15.1±2.8 mV, respectively (Figure S3). Thus, the favorable particle size and positive charge density of RGP nanoparticles made them suitable for achieving the cellular uptake in tumor cells. To further detect the reproducibility of this strategy, three batches of RGP nanoparticles were prepared, and then SDS-PAGE, hydrodynamic diameter and zeta potential analysis were conducted (Figures S4 and S5). Clearly, there were no distinct differences between these samples, indicating the good reproducibility of RGP nanoparticles.
To evaluate whether the genipin-mediated crosslinking reaction could influence the enzymatic activity, the activity of RGP was measured using a commercial RNase A activity detection kit. As shown in Figure 5, the fluorescence intensity of RNase A and RGP was observed to increase rapidly at an early stage and then become constant after 15 mins, which demonstrated the favorable enzymatic activity of RGP nanoparticles. Compared with free RNase A, the RGP activity was slightly reduced by ca. 27% but it still remained active. Simultaneously, the secondary structure of RNase A and RGP was determined by far UV circular dichroism spectra (Figure 6). Clearly, both RNase A and RGP exhibited typical α-helix conformation with two troughs at wavelengths at 209 and 222 nm. Based on the online analysis using software K2D2 (http://cbdm-01.zdv.uni-mainz.de/~andrade/k2d2/), the content of α-helix was identified to be decreased after genipin crosslinking (15.43% and 9.86% for RNase A and RGP, respectively). Meanwhile, the β-sheet content increased from 26.43% in RNase A to 41.03% in RGP nanoparticles. Thus, the change of secondary structure was probably a key reason for the slight deactivation of RNase A in the RGP group.
 |
Figure 5 The enzymatic activity of RNase A and RGP nanoparticles. |
 |
Figure 6 Circular dichroism spectra of RNase A and RGP. |
Owing to the introduction of cationic PEI25K, the hybrid system RGP is anticipated to be favorable for the interaction with cell membrane, thereby facilitating the cellular uptake. The delivery efficiency of RGP was evaluated through flow cytometry and CLSM, using HeLa cells as a model (Figure 7). Obviously, red fluorescence in HeLa cells could be detected owing to the intrinsic characteristic of genipin in the RGP sample, indicating the intracellular uptake of RNase A. Meanwhile, the phenomenon demonstrated that RGP nanoparticles could maintain the unique property of genipin after the crosslinking reaction. Generally, RNase A executed its antitumor function in the cytosol,35,36 but our results showed that some RGP nanoparticles exhibited a distribution in the nucleus. These results meant that the RGP nanoparticles could efficiently achieve the endosomal escape after the cellular uptake and even enter the nucleus. Thus, we conducted the endosomal escape analysis of RGP nanoparticles through CLSM images (Figure 8). At 2 hrs, obvious colocation of green and red fluorescence could be observed, implying the entrance in lysosomes of RGP nanoparticles. However, strong red fluorescence could be detected in the cytosol after 6 hrs, not merged with green fluorescence. Thus, it could be concluded that RGP nanoparticles could realize efficient endosomal escape owing to the “proton sponge” effect from PEI25K component. Moreover, flow cytometric analysis indicated that the fluorescence intensity of RGP exhibited an increasing tendency with the improvement of RNase A concentration in the system (Figure 7B and C). These results implied that the fluorescence intensity in the tumor cells increased in an RNase A dose-dependent manner. Notably, almost all the nanoparticles could be uptaken by the cells at an RNase A concentration of 8 μg/mL, with a delivery efficiency of 97.9%. Overall, the protein–polymer hybrid system RGP could realize an efficient intracellular delivery of therapeutic protein RNase A owing to the introduction of cationic carrier PEI25K, and thus it was likely to obtain ideal anti-tumor potency.
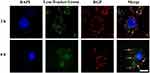 |
Figure 8 Endosomal escape analysis of RGP nanoparticles through CLSM. The scale bar is 50 μm. |
After the successful internalization of RGP, the in vitro antiproliferative effects of RNase A, PEI25K and RGP against HeLa cells were evaluated by MTT assay. In the present research, 0.1% Triton X-100 was employed as the positive control and the results are summarized in Figure S6. As shown in Figure 9, compared with PEI25K and RNase A, RGP nanoparticles exhibited obvious antiproliferative effects at an RNase A concentration of 4 and 5 μg/mL, owing to the successful intracellular delivery of RNase A and the further cleavage of RNA molecules in tumor cells. Meanwhile, RGP nanoparticles showed the antiproliferative effects in a dose-dependent manner, with only 43.9% of cell viability at an RNase A concentration of 5 μg/mL. Further, more dead cells could be observed in RGP group through live/dead staining assay (Figure 10), where the dead cells emitted red fluorescence after the staining with ethidium homodimer while the viable cells were labeled green due to the calcein acetoxymethyl ester (calcein AM) staining.37 Thus, all these results demonstrated the excellent antiproliferative effects of RGP nanoparticles owing to the successful delivery of RNase A with favorable antitumor response.
 |
Figure 10 Live/dead staining assay of HeLa cells after the treatment with RNase A and RGP. The scale bar is 200 μm. |
To further validate whether the proliferative inhibition was associated with the cell apoptosis, HeLa cells after RGP treatment was intuitively visualized through TUNEL staining. As shown in Figure 11, an obvious structural deformation of the nucleus and stronger green fluorescence could be simultaneously observed after the treatment with RGP for 48 hrs, indicating that RGP nanoparticles could efficiently induce the cell apoptosis. Moreover, the cell apoptosis was measured through flow cytometric analysis after cell staining with Annexin V-FITC and PI (Figure 12). In comparison to the control group, free RNase A did not induce any apoptosis, and only a slight apoptotic ratio could be detected for the PEI25K treatment which was probably relied on the intrinsic cytotoxicity of PEI25K. Remarkably, an obvious induction of cell apoptosis could be found for the RGP group with a total apoptotic ratio of ca. 46.2%. Thus, the antiproliferative effect triggered by RGP nanoparticles was attributed to the induction of cell apoptosis after the successful intracellular delivery of therapeutic protein RNase A.
 |
Figure 11 TUNEL assay of HeLa cells after the treatment with RNase A and RGP. The scale bar is 200 μm. |
 |
Figure 12 The cell apoptosis analysis of HeLa cells after the treatment with different samples through flow cytometry: (A) control; (B) RNase A; (C) PEI25K; and (D) RGP nanoparticles. |
Conclusion
In conclusion, therapeutic protein RNase A and cationic polymer PEI25K were covalently crosslinked using genipin as a crosslinker to construct a protein–polymer hybrid nanoparticle, aiming at the efficient intracellular delivery of RNase A. The RGP nanoparticle could be efficiently uptaken by cancer cells with a delivery efficiency of 97.9% and induce the apoptosis by cleaving the RNA molecules in the cytosol (total apoptotic ratio of 46.2%), thereby affording to the inhibition of cell proliferation. Additionally, the intrinsic red fluorescence of RGP from the introduction of genipin made it a potential theranostic delivery system for obtaining the combination of protein-based therapy and in vivo imaging. Overall, we believe that the genipin-mediated crosslinking technique will provide a promising tool for achieving the delivery of therapeutic proteins for the treatment of various diseases with improved therapeutic efficacy and minimized side effects.
Acknowledgment
The authors gratefully acknowledge the support from National Key R&D Program of China (2018YFC1105401), National Natural Science Foundation of China (81673502 and 81872928), Province-University Cooperation Project of Jilin Province (SXGJQY2017-4), Science and Technology Department of Jilin Province (20190201288JC), Education Department of Jilin Province (JJKH20190010KJ) and Graduate Innovation Program of Jilin University (10183201820).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. doi:10.1038/nrd2399
2. Yin L, Yuvienco C, Montclare JK. Protein based therapeutic delivery agents: contemporary developments and challenges. Biomaterials. 2017;134:91–116. doi:10.1016/j.biomaterials.2017.04.036
3. Pisal DS, Kosloski MP, Balu-Iyer SV. Delivery of therapeutic proteins. J Pharm Sci. 2010;99:2557–2575. doi:10.1002/jps.22054
4. Wang M, Zuris JA, Meng F, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci USA. 2016;113:2868–2873. doi:10.1073/pnas.1520244113
5. Leland PA, Raines RT. Cancer chemotherapy - ribonucleases to the rescue. Chem Biol. 2001;8:405–413.
6. Wang M, Alberti K, Sun S, et al. Combinatorially designed lipid-like nanoparticles for intracellular delivery of cytotoxic protein for cancer therapy. Angew Chem Int Ed. 2014;53:2893–2898. doi:10.1002/anie.201311245
7. Wang M, Sun S, Neufeld CI, et al. Reactive oxygen species-responsive protein modification and its intracellular delivery for targeted cancer therapy. Angew Chem Int Ed. 2014;53:13444–13448. doi:10.1002/anie.201407234
8. He H, Chen Y, Li Y, et al. Effective and selective anti-cancer protein delivery via all-functions-in-one nanocarriers coupled with visible light-responsive, reversible protein engineering. Adv Funct Mater. 2018;28:1706710. doi:10.1002/adfm.201706710
9. Liu M, Shen S, Wen D, et al. Hierarchical nanoassemblies-assisted combinational delivery of cytotoxic protein and antibiotic for cancer treatment. Nano Lett. 2018;18:2294–2303. doi:10.1021/acs.nanolett.7b04976
10. Shao D, Li M, Wang Z, et al. Bioinspired diselenide-bridged mesoporous silica nanoparticles for dual-responsive protein delivery. Adv Mater. 2018;30:1801198. doi:10.1002/adma.201801198
11. Akash MSH, Rehman K, Chen S. Natural and synthetic polymers as drug carriers for delivery of therapeutic proteins. Polym Rev. 2015;55:371–406. doi:10.1080/15583724.2014.995806
12. Rehman K, Akash MSH, Akhtar B, et al. Delivery of therapeutic proteins: challenges and strategies. Curr Drug Targets. 2016;17:1172–1188.
13. Chang J, Chen X, Glass Z, et al. Integrating combinatorial lipid nanoparticle and chemically modified protein for intracellular delivery and genome editing. Acc Chem Res. 2019;52:665–675. doi:10.1021/acs.accounts.8b00493
14. Zou S, Scarfo K, Nantz MH, et al. Lipid-mediated delivery of RNA is more efficient than delivery of DNA in non-dividing cells. Int J Pharm. 2010;389:232–243. doi:10.1016/j.ijpharm.2010.01.019
15. Liu X, Zhang P, He D, et al. pH-reversible cationic RNase A conjugates for enhanced cellular delivery and tumor cell killing. Biomacromolecules. 2015;17:173–182. doi:10.1021/acs.biomac.5b01289
16. D’Astolfo DS, Pagliero RJ, Pras A, et al. Efficient intracellular delivery of native proteins. Cell. 2015;161:674–690. doi:10.1016/j.cell.2015.03.028
17. Altinoglu SA, Wang M, Li KQ, et al. Intracellular aelivery of the PTEN protein using cationic lipidoids for cancer therapy. Biomater Sci. 2016;4:1773–1780. doi:10.1039/c6bm00580b
18. Jain A, Barve A, Zhao Z, et al. Comparison of avidin, neutravidin, and streptavidin as nanocarriers for efficient siRNA delivery. Mol Pharm. 2017;14:1517–1527. doi:10.1021/acs.molpharmaceut.6b00933
19. Tai W, Gao X. Functional peptides for siRNA delivery. Adv Drug Deliver Rev. 2017;110–111:157–168. doi:10.1016/j.addr.2016.08.004
20. Wang X, Li Y, Li Q, et al. Hyaluronic acid modification of RNase A and its intracellular delivery using lipid-like nanoparticles. J Control Release. 2017;263:39–45. doi:10.1016/j.jconrel.2017.01.037
21. Chen TT, Yi JT, Zhao YY, et al. Biomineralized metal-organic framework nanoparticles enable intracellular delivery and endo-lysosomal release of native active proteins. J Am Chem Soc. 2018;140:9912–9920. doi:10.1021/jacs.8b04457
22. Yu C, Tan E, Xu Y, et al. A guanidinium-rich polymer for effiecient cytosolic delivery of native proteins. Bioconjug Chem. 2019;30:413–417. doi:10.1021/acs.bioconjchem.8b00753
23. Zhang J, Wu D, Xing Z, et al. N-isopropylacrylamide-modified polyethylenimine-mediated p53 gene delivery to prevent the proliferation of cancer cells. Colloids Surf B Biointerfaces. 2015;129:54–62. doi:10.1016/j.colsurfb.2015.03.032
24. Liang S, Duan Y, Xing Z, et al. Inhibition of cell proliferation and migration by chondroitin sulfate-g-polyethylenimine-mediated miR-34a delivery. Colloids Surf B Biointerfaces. 2015;136:577–584. doi:10.1016/j.colsurfb.2015.09.054
25. Xing Z, Gao S, Duan Y, et al. Delivery of DNAzyme targeting aurora kinase A to inhibit the proliferation and migration of human prostate cancer. Int J Nanomed. 2015;10:5715–5727.
26. Ewe A, Przybylski S, Burkhardt J, et al. A novel tyrosine-modified low molecular weight polyethylenimine (P10Y) for efficient siRNA delivery in vitro and in vivo. J Control Release. 2016;230:13–25. doi:10.1016/j.jconrel.2016.03.034
27. Han H, Shi H, Wu D, et al. Genipin-cross-linked thermophilic histone-polyethylenimine as a hybrid gene carrier. ACS Macro Lett. 2015;4:575–578. doi:10.1021/acsmacrolett.5b00141
28. Lu H, Lu T, Chen C, Mi F-L. Development of genipin-crosslinked and fucoidan-adsorbed nano-hydroxyapatite/hydroxypropyl chitosan composite scaffolds for bone tissue engineering. Int J Biol Macromol. 2019;128:973–984. doi:10.1016/j.ijbiomac.2019.02.010
29. Silva NFN, Saint-Jalmes A, de Carvalho AF, et al. Development of casein microgels from cross-linking of casein micelles by genipin. Langmuir. 2014;30:10167–10175. doi:10.1021/la502274b
30. Xu J, Strandman S, Zhu JX, Barralet J, Cerruti M. Genipin-crosslinked catechol-chitosan mucoadhesive hydrogels for buccal drug delivery. Biomaterials. 2015;37:395–404. doi:10.1016/j.biomaterials.2014.10.024
31. Suh J, An Y, Tang BC, et al. Real-time gene delivery vector tracking in the endo-lysosomal pathway of live cells. Microsc Res Tech. 2012;75:691–697. doi:10.1002/jemt.21113
32. Shukla RS, Jain A, Zhao Z, Cheng K. Intracellular trafficking and exocytosis of a multi-component siRNA nanocomplex. Nanomedicine. 2016;12:1323–1334. doi:10.1016/j.nano.2016.02.003
33. Zhao Z, Li Y, Jain A, et al. Development of a peptide-modified siRNA nanocomplex for hepatic stellate cells. Nanomedicine. 2018;14:51–56. doi:10.1016/j.nano.2017.08.017
34. Gu X, Wei X, Fan Q, et al. cRGD-decorated biodegradable polytyrosine nanoparticles for robust encapsulation and targeted delivery of doxorubicin to colorectal cancer in vivo. J Control Release. 2019;301:110–118. doi:10.1016/j.jconrel.2019.03.005
35. Hu S, Chen X, Lei C, et al. Charge designable and tunable GFP as a target pH-responsive carrier for intracellular functional protein delivery and tracing. Chem Commun. 2018;54:7806–7809. doi:10.1039/C8CC03285H
36. Zhao S, Duan F, Liu S, et al. Efficient intracellular delivery of RNase A using DNA origami carriers. ACS Appl Mater Interfaces. 2019;11:11112–11118. doi:10.1021/acsami.8b21724
37. Wu D, Wang C, Yang J, et al. Improving the intracellular drug concentration in lung cancer treatment through the codelivery of doxorubicin and miR-519c mediated by porous PLGA microparticle. Mol Pharm. 2016;13:3925–3933. doi:10.1021/acs.molpharmaceut.6b00702
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.