Back to Journals » Drug Design, Development and Therapy » Volume 18
A Comprehensive Study to Investigate the Tumor-Suppressive Role of Radix Bupleuri on Gastric Cancer with Network Pharmacology and Molecular Docking
Authors Lv L, Du J, Wang D, Yan Z
Received 19 September 2023
Accepted for publication 16 January 2024
Published 8 February 2024 Volume 2024:18 Pages 375—394
DOI https://doi.org/10.2147/DDDT.S441126
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tuo Deng
Long Lv,1,* Jinghu Du,1,* Daorong Wang,2 Zeqiang Yan2
1Department of General Surgery, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Xiangyang, Hubei, 441021, People’s Republic of China; 2Department of Gastroenterology, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Xiangyang, Hubei, 441021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Daorong Wang; Zeqiang Yan, Department of Gastroenterology, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Xiangyang, Hubei, 441021, People’s Republic of China, Tel +13581425716 ; +13597505970, Email [email protected]; [email protected]
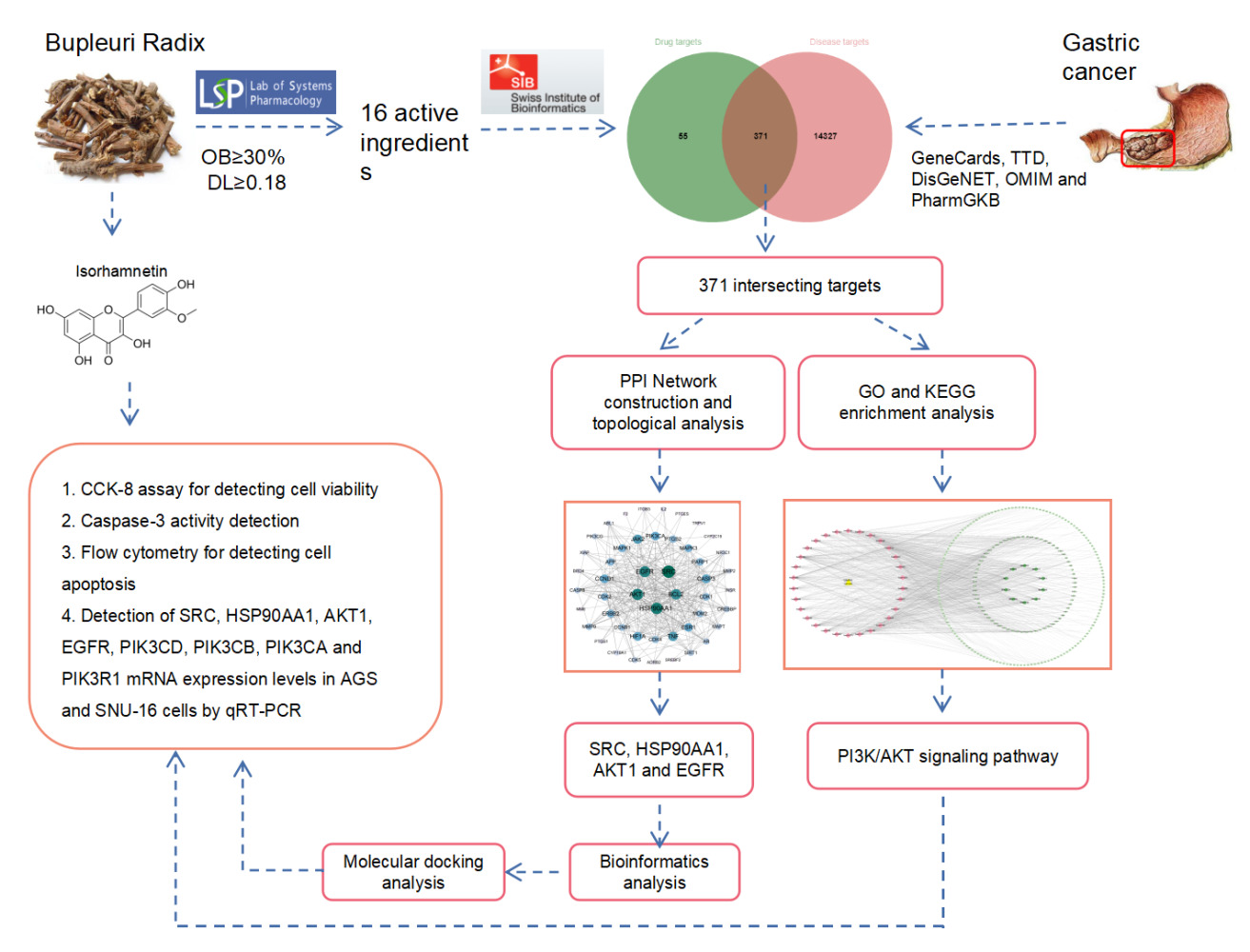
Background: Gastric cancer (GC) is a common fatal malignancy. The aim of this study was to explore and validate the tumor-suppressive role and mechanism of Radix Bupleuri in GC.
Methods: The active constituents of Radix Bupleuri were screened using TCMSP database. SwissTargetPrediction database was used to predict potential target genes of the compounds. GeneCards, TTD, DisGeNET, OMIM, and PharmGKB databases were used to search for GC-related targets. STRING database and Cytoscape 3.10 software were used for protein–protein interaction network construction and screening of core targets. DAVID database was used for GO and KEGG analyses. Core targets were validated using molecular docking. Cell proliferation and apoptosis were detected using CCK-8 and flow cytometry after GC cells were treated with isorhamnetin. The mRNA and protein expression levels of genes were detected using qRT PCR and Western blot. The metastasis potential of GC cells was evaluated in a nude mouse model.
Results: A total of 371 potential targets were retrieved by searching the intersection of Radix Bupleuri and GC targets. Petunidin, 3’,4’,5’,3,5,6,7-Heptamethoxyflavone, quercetin, kaempferol, and isorhamnetin were identified as the main bioactive compounds in Radix Bupleuri. SRC, HSP90AA1, AKT1, and EGFR, were core targets through which Radix Bupleuri suppressed GC. The tumor-suppressive effect of Radix Bupleuri on GC was mediated by multiple pathways, including PI3K-AKT, cAMP, and TNF signaling. The key compounds of Radix Bupleuri had good binding affinity with the core target. Isorhamnetin, a key component of Radix Bupleuri, could inhibit proliferation and metastasis, and induces apoptosis of GC cells. In addition, isorhamnetin could also reduce the mRNA expression of core targets, and the activation of PI3K/AKT pathway.
Conclusion: This study identified potential targets and pathways of Radix Bupleuri against GC through network pharmacology and molecular docking, providing new insights into the pharmacological mechanisms of Radix Bupleuri in GC treatment.
Keywords: gastric cancer, Radix Bupleuri, network pharmacology, molecular docking
Graphical Abstract:
Introduction
Gastric cancer (GC) is the third leading cause of cancer-related death worldwide; in 2020, 1,089,103 new cases of GC and 789,793 deaths were reported globally.1,2 More than half of the global GC cases occur in East Asia, with the majority of patients from China.3 Many factors, such as H. pylori infection, diet, smoking, obesity, and genetic changes, are related to the incidence of GC.4 Although advances in surgery, radiotherapy, chemotherapy, targeted therapy, and immunotherapy have significantly improved the survival rate of patients with GC, the prognosis of most patients with advanced GC remains disappointing.5,6 Therefore, it is necessary to explore novel, safe, and effective GC treatment strategies.
Traditional Chinese medicine has great potential in improving the quality of life of GC patients.7,8 Radix Bupleuri belongs to the Umbelliferae plants, and its dried root is widely used in traditional Chinese medicine, which has a medicinal history of more than 2000 years.9 In China, Radix Bupleuri is used to treat cold with fever, influenza, hepatitis, malaria, menopause, and hyperlipidemia, and is an important constituent of many compound preparations.9–11 Radix Bupleuri contains triterpenoid saponins (Radix Bupleuri saponins), polyacetylene, flavonoids, lignans, fatty acids, and sterols and exhibits a variety of pharmacological activities, including anti-inflammatory, antifungal, antiviral, antitumor, hepatoprotective, neuroprotective, and immunomodulatory effects.12 The active ingredient of Radix Bupleuri is found to inhibit the growth of triple-negative breast cancer cells by targeting β-catenin signaling;13 it also inhibits the malignancy of human neuroblastoma cells.14 Some biologically active components of Radix Bupleuri have been reported to suppress the malignancy of GC cells.15 However, the exact mechanism by which Radix Bupleuri exerts its tumor-suppressive properties during GC treatment remains unclear.
Network pharmacology, a comprehensive approach that combines systems biology and network informatics, has been widely used in the exploration and development of new drugs in recent years.16–18 In this study, we screened the bioactive ingredients of Radix Bupleuri for GC treatment and explored the downstream molecular mechanisms of action of Radix Bupleuri using molecular docking. Furthermore, we investigated the tumor-suppressive effects of isorhamnetin, a crucial component of Radix Bupleuri on the GC cells. This study provides a theoretical basis for the clinical treatment of GC using Radix Bupleuri.
Materials and Methods
Screening of Bioactive Compounds from Radix Bupleuri and Identification of Their Potential Target Genes
With Traditional Chinese Medicine System Pharmacology database (TCMSP database, http://lsp.nwu.edu.cn/tcmsp.php), according to the oral bioavailability (OB) 30% or higher, drug similarity (DL) of 0.8 or higher, potential effective bioactive ingredients of Radix Bupleuri were obtained. PubChem (https://pubchem.ncbi.nlm.nih.gov) and SwissTargetPrediction (http://www.swisstargetprediction.ch) databases were used to search for the target genes of these ingredients.
Screening of GC Related Targets
GC-related genes were obtained from the Human Genome Database (GeneCards, https://www.genecards.org/), Therapeutic Target Database (TDD, https://db.idrblab.net/ttd/), Online Human Mendel Database (OMIM, https://omim.org/), Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB, https://www.pharmgkb.org/), and Human Disease Gene Database (DisGeNET, https://www.disgenet.org/). After merging and deleting repeated genes, GC-related genes were identified.
Screening and Network Construction of Potential Targets of Radix Bupleuri for GC Treatment
Using the jvenn online platform (http://www.bioinformatics.com.cn/static/others/jvenn/example.html), a Venn diagram was generated to obtain the intersection of Radix Bupleuri target and GC-related genes. These genes are considered targets of Radix Bupleuri for GC treatment. Cytoscape 3.10 software was used to construct the bioactive ingredient – target gene network, and calculate the degree value of active ingredients.
Construction of Protein–Protein Interaction (PPI) Networks and Screening of Central Targets
Genes at the intersection of Radix Bupleuri target genes and GC-related genes were imported into the STRING database (https://cn.string-db.org/), and a PPI network diagram was constructed. The PPI network was visualized using Cytoscape 3.10 software and the Centiscape 2.2 plug-in was used to identify the central genes of Radix Bupleuri against GC.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analyses
Genes at the intersection of Radix Bupleuri target genes and GC-related genes were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/tools.jsp) for biological processes (BP), cellular components (CC), molecular functions (MF), and KEGG analysis. With P<0.01 as the screening condition, the key biological functions and signaling pathways of Radix Bupleuri against GC were predicted. Using the P value, the top 10 GO terms and top 30 KEGG pathways were visualized. Cytoscape 3.10 software was used to plot the top 30 KEGG pathway target graphs.
Molecular Docking
X-ray crystal structures of the proteins of core gene targets (SRC, HSP90AA1, AKT1, and EGFR) were obtained from the Protein Data Bank (PDB) database (https://www.rcsb.org/). From the PubChem database (https://www.ncbi.nlm.nih.gov/pccompound/), the 3D conformer figures of the top five key bioactive ingredients with the highest degrees were downloaded from the PubChem database (https://www.ncbi.nlm.nih.gov/pccompound/). With YinFu cloud computing platform (https://cloud.yinfotek.com/console/tools), SDF files were converted into mol2 format. Water molecules were removed using PyMOL software, and hydrogen atoms were added to the protein crystal structures. The mol2 file of the ingredients and PDB file of the protein crystal structure were then converted into a PDBQT format file using AutoDock Tools, and molecular docking was performed using the AutoDock Vina software (version 1.1.2). The binding energy with the highest score was selected for subsequent analysis, and the results were visualized using the PyMOL software.
Bioinformatics Analysis
UALCAN database (https://ualcan.path.uab.edu/analysis.html) was used to analyze the expression patterns of core targets (SRC, HSP90AA1, AKT1, and EGFR) in TCGA STAD cohort. The protein expression levels of the core targets were investigated using HPA database (https://www. Protein atlas). Kaplan-Meier Plotter database (http://kmplot.com/analysis/) was used for prognosis analysis. The Tumor Immune Estimation Resource (TIMER) online tool (https://cistrome.shinyapps.io/timer/) was used to study the association between the expression levels and somatic copy number changes of the core targets and the infiltration of different types of immune cells.
Cell Culture
Human GC cell lines AGS and SNU-16 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were placed in culture dishes containing Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), 100 U/mL penicillin and 100 μg/mL streptomycin in an incubator containing 5% CO2 at 37 °C.
Cell Viability Assay
Cell viability was measured using a cell counting Kit 8 (CCK-8, Beyotime, Shanghai, China). AGS and SNU-16 cells were inoculated into 96-well plates at a density of 0.5×104 cells/well. Different concentrations (0, 10, 20, 30, 40, 50, and 60 μM) of isorhamnetin were used (purity ≥95%, CAS No.17794; Sigma-Aldrich, St. Louis, MO, USA) to treat the cells for 24 or 48 h. Then, 10 μL of the CCK-8 solution was added to each well. After incubation at 37°C for 4h, the absorbance values of each well at 450 nm wavelength were detected by a microplate reader (Molecular Devices, Sunnyvale, CA, USA). An inhibition curve was subsequently drawn using GraphPad Prism software to calculate the 50% inhibitory concentration (IC50) of isorhamnetin on cell growth.
Evaluation of Caspase-3 Activity
A Caspase-3 activity detection kit (Abcam, Shanghai, China) was used to detect caspase-3 activity, according to the manufacturer’s protocol. After 24 h of treatment with 0 and 50 μM isorhamnetin, the cells were harvested and lysed at 4°C for 15 min using cell lysis buffer. The suspension was then centrifuged at 12,000 × g at 4°C for 10 min. The supernatant was then collected, and 10 µL of caspase-3 substrate Ac-LEHD-pNA and 10 µL of the supernatant were mixed and added to 80 µL of reaction buffer. After incubation at 37°C for 2 h, absorbance was measured at 405 nm wavelength using a microplate reader (Molecular Devices, Sunnyvale, CA, USA), and the relative caspase-3 activity was normalized to the control group.
Apoptosis Assay
The Annexin V-Fluorescein Isothiocyanate (FITC)/Propidium Iodide (PI) apoptosis detection kit (BD Pharmingen, San Diego, CA, USA) was used to detect apoptosis in the AGS and SNU-16 cells. Briefly, AGS and SNU-16 cells were seeded in a 6-well plate at a density of 5×105 cells/well and treated with 0 and 50 μM isorhamnetin for 24 h. The treated cells were collected, washed twice with cold phosphate buffer saline (PBS), and resuspended in binding buffer at a concentration of 1×106 cells/mL. The cells were then incubated with 5 μL Annexin V-FITC and 5 μL PI at room temperature in the dark for 15 min. After washing the cells with binding buffer, the apoptosis rates of AGS and SNU-16 cells were measured using a flow cytometer (BD Biosciences, San Jose, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from AGS and SNU-16 cells treated with different concentrations of isorhamnetin (0 and 50 μM) for 24h with TRIzol reagents (Invitrogen, Carlsbad, CA, USA). RNA concentration and purity were determined by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA (800 ng) from each group was reverse-transcribed into cDNA using the M-MLV Reverse Transcription Kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. SYBR Green PCR Master MIX kit (Vazyme, Nanjing, China) was used to prepare the PCR reaction system, and cDNA was amplified, and a Roche LightCycler 480 (Roche Diagnostics, Basel, Switzerland) was used to detect the expression level. GAPDH was used as the internal control, and the relative expression of the target gene was calculated by 2−ΔΔCt method. The primer sequences used in this study are as follows: SRC forward, 5’-GAGCGGCTCCAGATTGTCAA-3’, reverse, 5’-CTGGGGATGTAGCCTGTCTGT-3’; HSP90AA1 forward, 5’-AGGAGGTTGAGACGTTCGC-3’, reverse, 5’-AGAGTTCGATCTTGTTTGTTCGG-3’; AKT1 forward, 5’-CAAGGTGATCCTGGTGAA-3’, reverse, 5’-CGTGGGTCTGGAAAGAGT-3’; EGFR forward, 5’-CTTCGGGGAGCAGCGATgCGAC −3’, reverse, 5’-ACCAATACCTATTCCGTTACAC −3’; PIK3CD forward, 5’-TCAACTCACAGATCAGCCTC-3’, reverse, 5’-TTCACTTCTGGGTCGCACAA-3’; PIK3CB forward, 5’-TATTTGGACTTTGCGACAAGACT-3’, reverse, 5’-TCGAACGTACTGGTCTGGATAG-3’; PIK3R1 forward, 5’-AAGAAGTTGAACGAGTGGTTGG-3’, reverse, 5’-GCCCTGTTTACTGCTCTCCCC-3 ; GAPDH forward, 5’-TGACTTCAACAGCGACACCCA-3’, reverse, 5’-ACCCTGTTGCTGTAGCCAAA-3’.
Western Blot Assay
Total protein from AGS and SNU-16 cells was extracted using RIPA lysis buffer. Protein concentrations were quantified using a BCA assay kit (BioRad, Hercules, CA, USA). Equal amounts (30 µg) of proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). After being closed with 5% skimmed milk for 1h at room temperature, the membranes were incubated with primary antibodies: anti-phospho (p)-SRC (Y419) antibody (ab185617, 1:1000, Abcam), anti-SRC antibody (ab133283, 1:1000, Abcam), anti-p-EGFR (Y1068) antibody (ab40815, 1:1000, Abcam), anti-EGFR antibody (ab32077, 1:1000, Abcam), anti-p-AKT (T308) antibody (ab38449, 1:1000, Abcam), anti-AKT antibody (ab179463, 1:1000, Abcam), and anti-GAPDH antibody (ab181602, 1:1000, Abcam) at 4°C overnight. It was then incubated with HRP-coupled goat anti-rabbit secondary antibody (ab6721, 1:5000, Abcam) for 2h. The protein signal of the membrane was detected using the ECL Plus assay kit (Pierce, Rockford, IL, USA). Protein expression was quantified using ImageLab software version 4.1 (Bio-Rad Laboratories, Hercules, CA, USA).
Animal Experiments
The animal experiments were approved by the Ethics Committee of the Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science. Male BALB/c nude mice (4–6 weeks old) were purchased from Shulaibao Co., Ltd. To establish the lung/liver metastasis model, mice were divided into 2 groups (n = 10). After treatment with 50 μM isorhamnetin, AGS cells with the density of 4×105 were injected into the tail vein of each mouse. In the control group, AGS cells were treated with PBS. Mice were euthanized 4 weeks after injection. The lungs and livers were excised and fixed with 4% paraformaldehyde, paraffin-embedded, and stained with hematoxylin-eosin (H&E). Finally, the number of metastatic nodules in the lungs and liver was counted under a microscope to assess the severity of metastasis.
Statistical Analysis
SPSS software (version 20.0; SPSS Inc., Chicago, IL, USA) and GraphPad Prism 8 (Dotmatics, San Diego, CA, USA) were used for data processing and statistical analysis. The experiment was conducted in triplicate and independently repeated 3 times. All data are expressed as “mean ± standard deviation (SD)”. One-way analysis of variance (ANOVA) and Tukey’s post-hoc test were used for comparisons between the groups. Statistical significance was set at P <0.05.
Results
Screening Crucial Components of Radix Bupleuri and GC Related Genes
In TCMSP database, 16 active ingredients were identified based on OB ≥30% and DL ≥0.18, which includes 3’,4’,5’,3,5,6,7-Heptamethoxyflavone, petunidin, isorhamnetin, kaempferol, quercetin, areapillin, (+)-Anomalin, cubebin, linoleyl acetate, and saikosaponin C (Table 1 and Figure 1). Canonical SMILES corresponding to the 16 compounds was searched in the PubChem database and imported into Swiss Target Prediction database. Candidates with a probability >0 were selected for subsequent analysis. After deleting duplicate targets, 426 drug targets were identified. Meanwhile, 14,173 (relevance score > 1), 45, 312, 508 and 298 GC-related targets were selected from GeneCards, TTD, DisGeNET, OMIM and PharmGKB databases, respectively. The targets were merged, and duplicates were deleted, resulting in 14,698 targets (Figure 2A). We cross-analyzed 426 drug targets with 14,698 target diseases. The results identified 371 genes at the intersection as potential candidate target genes for Radix Bupleuri against GC (Figure 2B). The Radix Bupleuri component-target gene network was constructed using Cytoscape 3.10 software to further understand the correlation between Radix Bupleuri bioactive ingredients and GC-related genes. The network consisted of 389 nodes and 1398 edges (Figure 3A). The degree value of each node was calculated using the CytoNCA plug-in, and the greater the degree value, the greater the possibility that the compound plays a therapeutic role. Petunidin (degree = 92), 3’,4’,5’,3,5,6,7-Heptamethoxyflavone (degree value = 92), quercetin (degree value = 91), kaempferol (degree value = 91), and isorhamnetin (degree value = 91), with the highest degree values, were considered important bioactive compounds of Radix Bupleuri in the treatment of GC (Figure 3B).
 |
Table 1 Bioactive Components of Bupleuri Radix |
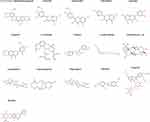 |
Figure 1 Chemical structure of 16 bioactive ingredients of Radix Bupleuri. |
Construction of PPI Network
The 371 genes mentioned above were imported into STRING database and set to a high confidence (0.7) for further analysis. The PPI network contained 371 nodes and 1665 edges (Figure 4A). The PPI network was visualized and analyzed using Cytoscape 3.10 software. All nodes were arranged in a circle according to the degree value; the larger the degree value, the darker the color and the more important they were in the PPI network (Figure 4B). Betweenness centrality ≥816.0949554896113, closeness centrality ≥0.000897228583357952, and degree centrality ≥9.869436201780415 were set as thresholds, and 48 core genes were finally obtained (Figure 4C). The nodes with the highest degree values included SRC (degree value = 67), HSP90AA1 (degree value = 61), AKT1 (degree value = 59), and EGFR (degree value = 57), which were considered key genes in the PPI network (Figure 4D). This suggests that Radix Bupleuri can treat GC via multiple bioactive ingredients that act on multiple targets.
GO and KEGG Analysis
We used DAVID database for GO and KEGG analyses to further clarify the function of Radix Bupleuri in GC. The results showed that 371 genes were significantly enriched in 459 biological processes (BP), 78 cell components (CC) and 143 molecular functions (MF) (P < 0.01). The top five BP terms were mainly enriched in protein phosphorylation (GO:0006468), response to xenobiotic stimulus (GO:0009410), protein autophosphorylation (GO:0046777), peptidyl-serine phosphorylation (GO:0018105), and negative regulation of the apoptotic process (GO:0043066). The top five terms of CC were integral components of the plasma membrane (GO:0005887), plasma membrane (GO:0005886), cytosol (GO:0005829), synapse (GO:0045202), and presynaptic membrane (GO:0099056). The top five MF terms were protein serine/threonine/tyrosine kinase activity (GO:0004712), protein kinase activity (GO:0004672), ATP-binding (GO:0005524), protein tyrosine kinase activity (GO:0004713), and protein serine/threonine kinase activity (GO:0004674). The top 10 BP, CC, and MF terms were selected for visualization (Figure 5A). KEGG analysis revealed 135 pathways related to the effects of Radix Bupleuri (P < 0.01). The first 30 KEGG pathways were visualized using bubble maps. As shown (Figure 5B), the target genes of Radix Bupleuri were mainly enriched in the PI3K-Akt signaling pathway (hsa04151), cAMP signaling pathway (hsa04024) and TNF signaling pathway (hsa04668), FoxO signaling pathway (hsa04068), and Phospholipase D signaling pathway (hsa04072). In addition, we classified and summarized the top 30 KEGG pathways and found that these KEGG pathways were mainly divided into four categories: Environmental Information Processing, Cellular Processes, Organismal Systems, and Human Diseases (Figure 5C). The results of the secondary classification of the KEGG pathway showed that the KEGG pathway was mainly enriched in cell growth and death and was also involved in biological systems, such as the endocrine, nervous, and immune systems (Figure 5D).
Construction of “Disease Target-Pathway” Network of GC Treated by Radix Bupleuri
Cytoscape 3.10 was used to visualize the top 30 KEGG pathways with the lowest P values and their corresponding targets, and a network with 258 nodes and 953 edges was obtained (Figure 6A). Analysis using the CytoNCA plug-in revealed that PIK3CD, PIK3CB, AKT1, PIK3CA, PIK3R1, MAPK1, MAPK3, MAP2K1, IKBKB, and CCND1 were significantly enriched in 30 KEGG pathways (Figure 6A). This suggests that the PI3K-AKT pathway may play a crucial role in Radix Bupleuri on GC. Subsequently, using KEGG PathView, the targets in the PI3K-AKT pathway were stained red (Figure 6B).
Molecular Docking
To further verify the downstream mechanism by which Radix Bupleuri suppresses the malignancy of GC cells, we investigated the possibility of binding between the core protein and the main bioactive compounds of Radix Bupleuri using molecular docking. It was revealed that, SRC (PDB ID:4M4Z), HSP90AA1 (PDB ID:4BQG), AKT1 (PDB ID:6HHG), EGFR (PDB ID:3POZ) had good binding activity with petunidin,3’,4’,5’,3,5,6,7-Heptamethoxyflavone, quercetin, kaempferol and isorhamnetin (Table 2). Furthermore, we visualized the molecular docking results using PyMol software (Figure 7).
 |
Table 2 The Binding Energy Between the Key Active Ingredients of Bupleuri Radix and the GC Core Targets |
The Expression Pattern of the Central Genes by Which of Radix Bupleuri Modulated GC
UALCAN database showed that the expression levels of SRC, HSP90AA1, AKT1, and EGFR were significantly higher in GC tissues than those in normal tissues (Figure 8A–D). In addition, it was found that high expression levels of SRC, HSP90AA1, AKT1, and EGFR were associated with late tumor stages in GC (Figure 8E–H). High expression levels of SRC, HSP90AA1, and AKT1 were also associated with lymph node metastasis in GC (Figure 8I–L). HPA database showed that the central target was expressed in both normal gastric and GC tissues. Notably, compared to normal gastric tissues, the expression levels of SRC and EGFR were increased in GC tissues, whereas the expression of AKT1 was decreased (Figure 9A). Kaplan-Meier Plotter database showed that high expression of SRC, AKT1, and EGFR was associated with shorter overall survival in GC patients (Figure 9B, P<0.05), suggesting that SRC, AKT1, and EGFR may be indicators of poor prognosis of GC patients. In addition, the relationships between somatic cell copy number changes in these genes and immune cell infiltration were analyzed using the TIMER website. The results showed that somatic copy number changes in SRC, HSP90AA1, AKT1, and EGFR significantly affected the proportions of B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells in the GC tissues (Figure 10A–D). SRC expression was significantly correlated with tumor purity (cor = 0.131; p = 1.04E-02), CD8+ T cells (cor = −0.113; p = 2.94e-02), macrophages (cor =−0.208; p=5.75e-05) 05), neutrophils (cor = −0.13; p = 1.20e-02), and dendritic cells (cor = −0.141; p=6.35e-03) (Figure 10E). HSP90AA1 expression negatively correlated with CD8+ T cells (cor = −0.194; p = 1.75e-04), CD4+ T cells (cor = −0.225; p = 1.38e-05), macrophages (cor =−0.261; p=3.56e-07) 05), neutrophils (cor =−0.134; p =9.91e-03), and dendritic cells (cor = −0.21; p=4.44e-05) (Figure 10F). AKT1 expression was correlated with CD8+ T cells (cor = −0.126; p = 1.50e-02), CD4+ T cells (cor = 0.104; p = 4.76e-02), and neutrophils (cor = −0.137; p =8.46e-03) (Figure 10G). EGFR expression was correlated with B cells (cor = 0.105; p = 4.31e-02), CD4+ T cells (cor = 0.143; p = 6.03e-03), and macrophages (cor = 0.115; p=2.74e-02) (Figure 10H). These findings suggest that these genes play a key role in regulating the tumor immune microenvironment in patients with GC, and Radix Bupleuri may modulate GC progression via these targets.
Isorhamnetin Inhibits the Malignancy of GC Cells
To verify the anticancer function of Radix Bupleuri in GC, we selected isorhamnetin, a key component of Radix Bupleuri, and investigated its tumor-suppressive role in the malignancy of GC cells. As shown (Figure 11A), isorhamnetin significantly reduced the viability of AGS and SNU-16 cells in a concentration- and time-dependent manner compared with that in the control group. IC50 values of AGS cells treated with isorhamnetin for 24 h and 48 h were 52.67 μM and 43.46 μM, respectively, and those of SNU-16 cells were 52.82 μM and 40.96 μM, respectively (Figure 11B). In the subsequent experiment, 50 μM isorhamnetin was selected as the concentration, and the cells were treated for 24 h. To investigate the effect of isorhamnetin on GC cell apoptosis, caspase-3 activity in GC cells was examined after pretreatment with isorhamnetin. Isorhamnetin significantly enhanced caspase-3 activity in AGS and SNU-16 cells (Figure 11C). Flow cytometry analysis showed that the apoptosis rates of AGS and SNU-16 cells pretreated with isorhamnetin were significantly higher than those in the control groups (Figure 11D). In addition, we verified the effect of isorhamnetin on the mRNA expression levels of central targets such as SRC, HSP90AA1, AKT1, and EGFR by qRT-PCR. The results showed that isorhamnetin reduced the mRNA expression of SRC, HSP90AA1, AKT1, and EGFR (Figure 11E). Compared to the control group, isorhamnetin also significantly inhibited the mRNA expression levels of PIK3CD, PIK3CB, PIK3CA, and PIK3R1 in AGS and SNU-16 cells (Figure 11F). Western blot showed that isorhamnetin significantly reduced the protein expression levels of p-SRC, p-EGFR, and p-AKT in AGS and SNU-16 cells (Figure 11H and I). These results showed that isorhamnetin, one of the main bioactive compounds in Radix Bupleuri, could repress the expression and activation of multiple oncogenes in GC cells. To further validate the effect of isorhamnetin on the malignancy of GC cells, a mouse models of lung and liver metastasis was constructed by injecting AGS cells into the tail vein. The results showed that compared with the control group, isorhamnetin treatment significantly reduced the number of metastatic lesions in the lungs and liver of mice (Figure 12A and B).
Discussion
When applied to treat human malignancies, traditional Chinese medicine has the characteristics of “multi-component, multi-target and multi-pathway”.19,20 Several studies have shown that some bioactive components of Radix Bupleuri, such as saikosaponin A, saikosaponin b2, and saikosaponin D, have anticancer activity against various tumors, such as neuroblastoma,14 liver cancer,21 breast cancer13,22 and GC.15 This study combined network pharmacology, molecular docking, in vitro experiments, and in vivo experiments to explore the potential mechanism of Radix Bupleuri in GC treatment, and preliminarily revealed the characteristics of ‘multi-component, multi-target and multi-pathway’ of Radix Bupleuri to repress GC progression.
In the present study, we found that Radix Bupleuri contains at least 16 bioactive ingredients, corresponding to 426 targets, and 371 potential targets were associated with GC tumorigenesis and progression. By constructing the component-target gene network, five key bioactive ingredients of Radix Bupleuri against GC were identified: petunidin,3’,4’,5’,3,5,6,7-Heptamethoxyflavone, quercetin, kaempferol, and isorhamnetin. Quercetin is a flavonoid widely found in edible fruits and vegetables and has anti-inflammatory, antiviral, and antitumor effects.23 Quercetin inhibits GC cell proliferation by activating the pyroptosis pathway.24 Kaempferol is a natural flavonoid and histone deacetylase (HDAC) inhibitor that has anticancer properties against a variety of human cancers, including GC.25 Kaempferol induces autophagic cell death in GC cells via the IRE1-JNK-CHOP pathway.26 Isorhamnetin inhibits proliferation of human GC cells through mitochondria-dependent apoptosis.27 Overall, the anticancer properties of these components may provide an important theoretical basis for the treatment of GC using Radix Bupleuri.
The PPI network showed that SRC, HSP90AA1, AKT1, and EGFR were the core targets of Radix Bupleuri for GC treatment. SRC is one of the most well-studied oncoproteins, and its high expression and enhanced activity are associated with poor survival in patients with GC.28 HSP90AA1, also known as HSP90α, is an isomer of the chaperone HSP90, and high expression of HSP90AA1 is associated with disease progression and poor survival in GC patients.29 AKT1, a serine/threonine-specific protein kinase, is an important component of the PI3K/AKT pathway and plays a crucial role in the regulation of cell proliferation, differentiation, invasion, apoptosis, glucose metabolism, and other biological processes.30 Abnormal high expression of AKT1 in GC tissues promotes proliferation of GC cells and modulates apoptosis.31 EGFR is a transmembrane glycoprotein and member of the receptor tyrosine kinase ErbB family.32 EGFR is highly expressed in GC and is a biomarker of poor prognosis, and is one of the most important regulators of cancer cell proliferation.33 These previous reports suggest that these core genes play an important role in the tumorigenesis and progression of GC and can be potential targets for Radix Bupleuri in GC treatment. Consistently, we found that SRC, HSP90AA1, AKT1, and EGFR were highly expressed in the GC tissues. High expression of SRC, AKT1, and EGFR is also associated with shorter overall survival in patients with GC. In addition, the expression of SRC, HSP90AA1, AKT1, and EGFR is associated with abnormal immune cell infiltration in GC. Importantly, this study confirmed that the key components of Bubularum petunidin,3’,4’,5’,3,5,6,7-Heptamethoxyflavone, quercetin, kaempferol, and isorhamnetin, had good binding affinities with SRC, HSP90AA1, AKT1, and EGFR, and isorhamnetin blocked the activation of SRC, AKT1 and EGFR, suggesting that Radix Bupleuri may modulate the immune microenvironment of GC tissues to block GC progression via these targets.
KEGG analysis showed that the PI3K/AKT signaling pathway may be the key pathway for the anti-GC effect of Radix Bupleuri. In this study, isorhamnetin, a key component of Radix Bupleuri, was found to inhibit the malignancy of GC cells in vitro and in vivo. The results of this study showed that isorhamnetin could improve the activity of caspase-3 in GC cells and effectively induce apoptosis. PI3K/AKT pathway is a negative regulator of apoptosis, and its overactivation can promote proliferation of GC cells and inhibit apoptosis.34,35 Therefore, the inhibition of the PI3K/AKT pathway may be a potential mechanism for GC therapy. In this study, we found that isorhamnetin reduced the mRNA levels of SRC, HSP90AA1, AKT1, EGFR, PIK3CD, PIK3CB, PIK3CA, and PIK3R1, which are all associated with PI3K/AKT activation in GC cells. This suggested that isorhamnetin inhibited GC cell proliferation and promoted apoptosis by regulating PI3K/AKT pathway.
This study has some limitations. First, public databases are updated in real-time, so the results of this study only partially reveal the mechanism of Radix Bupleuri in treating GC. Second, although isorhamnetin is the main active ingredient of Radix Bupleuri, it is still not fully representative of Radix Bupleuri, and the effects of other active ingredients need to be verified in the future. In addition, besides the PI3K-Akt signaling pathway, Radix Bupleuri has also been found to be involved in modulating other pathways, such as the TNF signaling pathway and FoxO signaling pathway. In the future, other mechanisms of action of Radix Bupleuri against GC need to be further investigated. Last but not least, the current study lacks the direct evidence to validate the interaction between the ingredients and the crucial targets, and necessary technologies such as HPLC-MS will help solve this problem in the following work.
In summary, this study elucidates the potential mechanism of action of Radix Bupleuri against GC. This study suggested that Radix Bupleuri blocks GC progression by acting on multiple targets and pathways. Our findings provide important information regarding the role of Radix Bupleuri in GC treatment and suggest that Radix Bupleuri is a promising candidate drug for clinical treatment of GC, and of course its safety and efficacy require more data from clinical trials, which are needed to be performed in the future.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Statement
This study was reviewed and approved by the Animal Ethics Committee of Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Wang J, Wang Y, Wang J, et al. DEAD-box helicase 56 functions as an oncogene promote cell proliferation and invasion in gastric cancer via the FOXO1/p21 Cip1/c-Myc signaling pathway. Bioengineered. 2022;13(5):13970–13985. doi:10.1080/21655979.2022.2084235
3. Zhang T, Chen H, Yin X, et al. Changing trends of disease burden of gastric cancer in China from 1990 to 2019 and its predictions: findings from global burden of disease study. Chin J Cancer Res. 2021;33(1):11–26. doi:10.21147/j.issn.1000-9604.2021.01.02
4. Machlowska J, Kapusta P, Baj J, et al. High-throughput sequencing of gastric cancer patients: unravelling genetic predispositions towards an early-onset subtype. Cancers. 2020;12(7):1981. doi:10.3390/cancers12071981
5. Gao C, Liu F, Ye Q, Guo A. Cancer-associated fibroblasts affect tumor metabolism and immune microenvironment in gastric cancer and identification of its characteristic genes. J Oncol. 2023;2023:1424589. doi:10.1155/2023/1424589
6. Le X, Mu J, Peng W, et al. DNA methylation downregulated ZDHHC1 suppresses tumor growth by altering cellular metabolism and inducing oxidative/ER stress-mediated apoptosis and pyroptosis. Theranostics. 2020;10(21):9495–9511. doi:10.7150/thno.45631
7. Huang Z, Wei P. Compound Kushen Injection for gastric cancer: a protocol of systematic review and meta-analysis. Medicine. 2019;98(45):e17927. doi:10.1097/MD.0000000000017927
8. Song S, Zhou J, Li Y, Liu J, Li J, Shu P. Network pharmacology and experimental verification based research into the effect and mechanism of aucklandiae radix-amomi fructus against gastric cancer. Sci Rep. 2022;12(1):9401. doi:10.1038/s41598-022-13223-z
9. Li X, Li X, Huang N, Liu R, Sun R. A comprehensive review and perspectives on pharmacology and toxicology of saikosaponins. Phytomedicine. 2018;50:73–87. doi:10.1016/j.phymed.2018.09.174
10. Yan LJ, Wang ZJ, Fang M, et al. Bupleuri radix for acute uncomplicated respiratory tract infection: a systematic review of randomized controlled trials. Front Pharmacol. 2022;12:787084. doi:10.3389/fphar.2021.787084
11. Chen Y, Wang J, Yuan L, Zhou L, Jia X, Tan X. Interaction of the main components from the traditional Chinese drug pair Chaihu-Shaoyao based on rat intestinal absorption. Molecules. 2011;16(11):9600–9610. doi:10.3390/molecules16119600
12. Yang F, Dong X, Yin X, Wang W, You L, Ni J. Radix bupleuri: a review of traditional uses, botany, phytochemistry, pharmacology, and toxicology. Biomed Res Int. 2017;2017:7597596. doi:10.1155/2017/7597596
13. Wang J, Qi H, Zhang X, et al. Saikosaponin D from Radix Bupleuri suppresses triple-negative breast cancer cell growth by targeting β-catenin signaling. Biomed Pharmacother. 2018;108:724–733. doi:10.1016/j.biopha.2018.09.038
14. Cheng T, Ying M. Antitumor effect of saikosaponin A on human neuroblastoma cells. Biomed Res Int. 2021;2021:5845554. doi:10.1155/2021/5845554
15. Hu J, Li P, Shi B, Tie J. Effects and mechanisms of saikosaponin D improving the sensitivity of human gastric cancer cells to cisplatin. ACS Omega. 2021;6(29):18745–18755. doi:10.1021/acsomega.1c01795
16. Shang L, Wang Y, Li J, et al. Mechanism of Sijunzi Decoction in the treatment of colorectal cancer based on network pharmacology and experimental validation. J Ethnopharmacol. 2023;302(Pt A):115876. doi:10.1016/j.jep.2022.115876
17. Liu J, Liu J, Tong X, et al. Network pharmacology prediction and molecular docking-based strategy to discover the potential pharmacological mechanism of Huai Hua San Against ulcerative colitis. Drug Des Devel Ther. 2021;15:3255–3276. doi:10.2147/DDDT.S319786
18. Gao J, Yang S, Xie G, Pan J, Zhu F. Integrating network pharmacology and experimental verification to explore the pharmacological mechanisms of aloin against gastric cancer. Drug Des Devel Ther. 2022;16:1947–1961. doi:10.2147/DDDT.S360790
19. Wang Y, Chu F, Lin J, et al. the main active ingredient of Dendrobium chrysotoxum Lindl, inhibits precancerous lesions of gastric cancer (PLGC) through suppression of the HRAS-PI3K-AKT signaling pathway as revealed by network pharmacology and in vitro experimental verification. J Ethnopharmacol. 2021;279:114399. doi:10.1016/j.jep.2021.114399
20. Han L, Han Y. Network pharmacology-based study on the active component and mechanism of the anti-gastric-cancer effect of herba sarcandrae. J Healthc Eng. 2021;2021:3001131. doi:10.1155/2021/3001131
21. You M, Fu J, Lv X, Wang L, Wang H, Li R. Saikosaponin b2 inhibits tumor angiogenesis in liver cancer via down‑regulation of VEGF/ERK/HIF‑1α signaling. Oncol Rep. 2023;50(1):136. doi:10.3892/or.2023.8573
22. Ma Q, Gao FF, He X, et al. Antitumor effects of saikosaponin b2 on breast cancer cell proliferation and migration. Mol Med Rep. 2019;20(2):1943–1951. doi:10.3892/mmr.2019.10385
23. Shen X, Si Y, Wang Z, Wang J, Guo Y, Zhang X. Quercetin inhibits the growth of human gastric cancer stem cells by inducing mitochondrial-dependent apoptosis through the inhibition of PI3K/Akt signaling. Int J Mol Med. 2016;38(2):619–626. doi:10.3892/ijmm.2016.2625
24. Rong Y, Liu SH, Tang MZ, Yang XJ. Quercetin inhibits the proliferative effect of gastric cancer cells by activating the pyroptosis pathway. Asian J Surg. 2023;S1015–9584(23):1063.
25. Yang L, Li H, Yang M, et al. Exploration in the mechanism of kaempferol for the treatment of gastric cancer based on network pharmacology. Biomed Res Int. 2020;2020:5891016. doi:10.1155/2020/5891016
26. Kim TW, Lee SY, Kim M, Cheon C, Ko SG. Kaempferol induces autophagic cell death via IRE1-JNK-CHOP pathway and inhibition of G9a in gastric cancer cells. Cell Death Dis. 2018;9(9):875. doi:10.1038/s41419-018-0930-1
27. Li Y, Fan B, Pu N, et al. Isorhamnetin suppresses human gastric cancer cell proliferation through mitochondria-dependent apoptosis. Molecules. 2022;27(16):5191. doi:10.3390/molecules27165191
28. Caner A, Asik E, Ozpolat B. SRC Signaling in Cancer and Tumor Microenvironment. Adv Exp Med Biol. 2021;1270:57–71.
29. Lee HW, Kim KM. Clinical significance of heat shock protein 90α expression as a biomarker of prognosis in patients with gastric cancer. Niger J Clin Pract. 2019;22(12):1698–1705. doi:10.4103/njcp.njcp_68_19
30. Deng T, Shen P, Li A, et al. CCDC65 as a new potential tumor suppressor induced by metformin inhibits activation of AKT1 via ubiquitination of ENO1 in gastric cancer. Theranostics. 2021;11(16):8112–8128. doi:10.7150/thno.54961
31. Liu HZ, Shan TD, Han Y, Liu XS. Silencing long non-coding RNA CASC9 inhibits colorectal cancer cell proliferation by acting as a competing endogenous RNA of miR-576-5p to regulate AKT3. Cell Death Discov. 2020;6(1):115. doi:10.1038/s41420-020-00352-5
32. Yu J, Feng H, Sang Q, et al. VPS35 promotes cell proliferation via EGFR recycling and enhances EGFR inhibitors response in gastric cancer. E Bio Medicine. 2023;89:104451.
33. Higaki E, Kuwata T, Nagatsuma AK, et al. Gene copy number gain of EGFR is a poor prognostic biomarker in gastric cancer: evaluation of 855 patients with bright-field dual in situ hybridization (DISH) method. Gastric Cancer. 2016;19(1):63–73. doi:10.1007/s10120-014-0449-9
34. Fattahi S, Amjadi-Moheb F, Tabaripour R, Ashrafi GH, Akhavan-Niaki H. PI3K/AKT/mTOR signaling in gastric cancer: epigenetics and beyond. Life Sci. 2020;262:118513. doi:10.1016/j.lfs.2020.118513
35. Rong L, Li Z, Leng X, et al. Salidroside induces apoptosis and protective autophagy in human gastric cancer AGS cells through the PI3K/Akt/mTOR pathway. Biomed Pharmacother. 2020;122:109726. doi:10.1016/j.biopha.2019.109726
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.