Back to Journals » Cancer Management and Research » Volume 10
A comparison of QuantStudio™ 3D Digital PCR and ARMS-PCR for measuring plasma EGFR T790M mutations of NSCLC patients
Authors Feng Q, Gai F, Sang Y, Zhang J, Wang P, Wang Y, Liu B, Lin D, Yu Y, Fang J
Received 2 August 2017
Accepted for publication 31 October 2017
Published 18 January 2018 Volume 2018:10 Pages 115—121
DOI https://doi.org/10.2147/CMAR.S148134
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Qin Feng,1,* Fei Gai,2,* Yaxiong Sang,2 Jie Zhang,3 Ping Wang,1 Yue Wang,1 Bing Liu,2 Dongmei Lin,1 Yang Yu,2 Jian Fang3
1Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Pathology, Peking University Cancer Hospital & Institute, 2Oncology Business Division, Beijing Novogene Bioinformatics Technology Co., Ltd, 3Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Department of Thoracic Oncology II, Peking University Cancer Hospital & Institute, Beijing, China
*These authors contributed equally to this work
Background: The AURA3 clinical trial has shown that advanced non-small cell lung cancer (NSCLC) patients with EGFR T790M mutations in circulating tumor DNA (ctDNA) could benefit from osimertinib.
Purpose: The aim of this study was to assess the usefulness of QuantStudio™ 3D Digital PCR System platform for the detection of plasma EGFR T790M mutations in NSCLC patients, and compare the performances of 3D Digital PCR and ARMS-PCR.
Patients and methods: A total of 119 Chinese patients were enrolled in this study. Mutant allele frequency of plasma EGFR T790M was detected by 3D Digital PCR, then 25 selected samples were verified by ARMS-PCR and four of them were verified by next generation sequencing (NGS).
Results: In total, 52.94% (69/119) had EGFR T790M mutations detected by 3D Digital PCR. In 69 positive samples, the median mutant allele frequency (AF) was 1.09% and three cases presented low concentration (AF <0.1%). Limited by the amount of plasma DNA, 17 samples (AF <2.5%) and eight samples (T790M-) were selected for verification by ARMS-PCR. Four of those samples were verified by NGS as a third verification method. Among the selected 17 positive cases, ten samples presented mutant allele frequency <0.5%, and seven samples presented intermediate mutant allele frequency (0.5%<AF<2.5%). However, only three samples (3/17) were identified as positive by ARMS-PCR, namely, P6 (AF =1.09%), P7 (AF =2.09%), and P8 (AF =2.21%). It is worth mentioning that sample P9 (AF =2.05%, analyzed by 3D Digital PCR) was identified as T790M- by ARMS-PCR. Four samples were identified as T790M+ by both NGS and 3D Digital PCR, and typically three samples (3/4) presented at a low ratio (AF <0.5%).
Conclusion: Our study demonstrated that 3D Digital PCR is a novel method with high sensitivity and specificity to detect EGFR T790M mutation in plasma.
Keywords: 3D Digital PCR, allele frequency, EGFR TKIs, resistance, osimertinib, erlotinib, gefitinib, icotinib
Introduction
Data from population-based registries collected by the National Central Cancer Registry of China (2009–2011) showed that lung cancer has the highest incidence and is the leading cause of cancer death in China. The results indicated that an estimated 733,300 new lung cancer cases and 610,200 lung cancer deaths would occur in China in 2015.1,2 Generally, non-small cell lung cancer (NSCLC) and small cell lung cancer are the major pathology subtypes of lung cancer.3 NSCLC comprises almost 80%–85% of all lung cancer cases.3,4 It is reported that activating mutation of EGFR could control cellular proliferation and survival of cancer cells.5 Point mutations of exon 21 (Leu858Arg) and exon 19 (in-frame) deletions are the most common activating mutations of EGFR, which constitutes more than 90% of known activating EGFR mutations.6 The frequency of EGFR mutation among Caucasian NSCLC populations is approximately 20%, compared with 60%~70% in never-smoking, Asian, adenocarcinoma lung cancer patients.7 EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib,8,9 gefitinib,10,11 afatinib12,13 or icotinib,14 have led to dramatic improvement in progression-free survival (PFS) compared to standard chemotherapy for lung cancer patients with specific EGFR mutations (p.L858R or exon 19 deletions).
Although the superiority of first-line gefitinib and erlotinib (the “first generation” EGFR TKIs) over standard chemotherapy has been established in several clinical trials, most patients who had responded to EGFR TKIs acquired resistance to them and had disease progression within 10~16 months.15 Of those resistance cases, 50%~65% developed a second EGFR mutation, T790M. This mutation is a critical amino acid that lies within the ATP-binding pocket of EGFR, when it occurs in cis with the original drug-sensitive EGFR mutation, it could reduce drug affinity of the kinase domain compared with cellular ATP.8,16–18 Neither the “first generation” EGFR TKIs nor the “second generation” EGFR TKIs (neratinib, afatinib, and dacomitinib) could overcome the resistance to EGFR TKIs, which is caused by the EGFR T790M mutation.19 The “third generation” EGFR TKIs (osimertinib, CO-1682, HM61713 and AC0010) have been demonstrated to irreversibly bind to the sensitive mutation form of EGFR (L858R, delE746-A750) and the gatekeeper mutation (T790M).20–22 Specifically, osimertinib has been approved by the US Food and Drug Administration (FDA) in 2015 for NSCLC patients with EGFR T790M mutation,23 and by the China Food and Drug Administration (CFDA) in 2017. The Phase III trial, AURA3, recruited 419 patients with T790M+ advanced NSCLC, the median duration of PFS with osimertinib was 10.1 months, 5.7 months longer than with platinum therapy plus pemetrexed.24
Recently, ctDNA has emerged as a specific and sensitive biomarker for EGFR variants’ detection.25,26 Minimal invasion is the most prominent advantage for ctDNA test. Another advantage, comprehensive analysis of the tumor heterogeneity, makes ctDNA test more popular for tumor research.27–29 However, the abundance of ctDNA varies from 0.01% to 67% for patients with different kinds of cancers or progression stages.26,30,31 Thus, there is an urgent need for a standardized, sensitive method for ctDNA testing. The non-digital platforms (ARMS and Cobas®) and the digital platforms (BEAMing digital PCR, Droplet digital PCR and next generation sequencing [NGS]-based methods) have been commonly used for analyzing the EGFR mutations in plasma ctDNA.32,33 AURA I reported that sensitivity of EGFR T790M plasma detection (by BEAMing) was 70%, compared with tissue genotyping (by Cobas). Approximately 45% false-negative plasma genotyping patients have significant outcomes (median PFS, 9.3 months) with osimertinib, according to T790M+ tissue genotyping.34 In this study, we introduced a super sensitive ctDNA T790M detection method through QuantStudio™ 3D Digital PCR System platform, which can detect low levels of somatic T790M alterations in plasma of patients. According to 3D digital PCR approach, the reaction is divided into 20,000 individual partitions, and the absolute copy number is calculated based on statistical interpretation of the number of partitions where the target mutation alleles have been detected, compared to those where wild-type alleles have been detected. It also gives the chance to detect T790M before EGFR TKI exposure, providing more accurate molecular diagnostic information to advanced NSCLC patients.
Methods
Experimental design
Plasma samples were collected from 119 NSCLC patients during January 2015 to September 2016, separated from 8–10 mL anticoagulant venous blood. All EGFR T790M mutations were detected using QuantStudio™ 3D Digital PCR System from plasma DNA. Then we selected T790M frequency <2.5% and partial T790M- samples, which were used to do ARMS-PCR single blind verification. Limited by the remaining amount of ctDNA, we only verified 25 samples; including four samples which were verified by NGS at the same time. The sensitivity and specificity of plasma EGFR T790M mutation genotyping by 3D Digital PCR were analyzed. This study was approved by the Medical Ethics Committee of Peking University Cancer Hospital and performed according to the Declaration of Helsinki principles. All patients signed informed consent for their samples to be used in a future study. All clinical data and samples were received anonymously.
ctDNA extraction
Peripheral blood samples were collected into BCT-EDTA tubes (Streck, Omaha, NE, USA). Plasma was isolated from blood by centrifuging at 1,600 g for 10 minutes at 4°C. Then the plasma was further centrifuged at 16,000 g for 10 minutes at 4°C, and was then stored at 80°C until ctDNA extraction. Plasma ctDNA was extracted from 2 mL of plasma from each patient with the QIAamp Circulating Nucleic Acid kit (Qiagen NV, Venlo, the Netherlands), following the manufacturer’s instructions.
Plasma ctDNA EGFR T790M detection
We used three methods to detect plasma ctDNA T790M in this research, 3D Digital PCR, NGS, and ARMS-PCR. We purchased all 3D Digital PCR reagents from Thermo Fisher Scientific (Waltham, MA, USA), custom ordered primers (T790M-F: 5′-GCATCTGCCTCACCTCCACC; T790M-R: 5′-ACCAGTTGAGCAGGTACTGGGAGC) and probes (T790M-P1(FAM):5′-AGCTCATCATGCAGCTCAT; WT-P2(VIC): 5′-AGCTCATCACGCAGCTCAT) from Life Technologies (Thermo Fisher Scientific), and performed the 3D Digital PCR analysis on a QuantStudio™ 3D Digital PCR System. We optimized the experimental process, the lowest detection rate of plasma T790M is 0.04%, stable detection frequency of plasma T790M is 0.1%, as previously published.35 The final 15 μL of TaqMan PCR reaction mixture was made up in the following way: 7.5 μL 2× QuantStudio™ 3D Digital PCR Master Mix, 0.75 μL 20× TaqManAssay (primer/probe mix), 6.75 μL diluted DNA (25 ng), and then loaded into the QuantStudio™ 3D Digital PCR Chip, which has 20000 mini-chambers. To perform the PCR using the ProFlex™ 2× Flat PCR System, thermal cycling profile was 10 minutes of incubation at 96°C, followed by 39 cycles of 56°C for 2 minutes, 98°C for 30 seconds, 56°C for 2 minutes, and then 4°C hold. We used the QuantStudio™ 3D Digital PCR instrument to read the chip. The subsequent analysis was performed with the QuantStudio 3D Analysis Suite Software.
Plasma ctDNA was sequenced using paired-end strategy on an Illumina HiSeq platform (Illumina, San Diego, CA, USA), as previously published.36 A KAPA Hyper Prep kit (Kapa Biosystems Inc., MA, USA) was used to construct the library. Then the prepped libraries were hybridized with SureSelectQXT Target Enrichment System (Agilent Technologies) to capture the targeted sequences. The concentration of each library was quantified using a QPCR NGS library quantification kit (Agilent Technologies, Santa Clara, CA, USA). After sequencing, the raw data were analyzed by self-developed analytical software. Every somatic mutation identified in ctDNA was checked by Integrative Genomics Viewer software and Samtools software.
ADx-ARMS (amplification refractory mutation system) EGFR mutation kit (Amoy Diagnostics, Xiamen, China) was used, and all experiments and genotype calling were performed following the manufacturer’s instructions.
Statistical analysis
The statistical data were analyzed with SAS software version 9.2 (SAS Institute Inc., Cary, NC, USA). The McNemar’s test and Fisher’s exact test were used to compare the consistency of plasma T790M mutation result between 3D digital PCR and ARMS-PCR.
Statistical figures were done with GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Clinico-morphological characteristics of patients
Peripheral blood samples obtained from 119 NSCLC patients, aged between 31 and 91 were tested in this study. Patients’ clinical characteristics are listed in Table 1. Sixty females and 59 males participated in this study cohort, of whom 82 were diagnosed with stage III–IV NSCLC. A total of 106 of 119 patients (89%) had adenocarcinoma or mixed histology. Treatment experienced patients accounted for 86.6% (103 of 119) of the total patients, and 92.2% (95 of 103) of them received EGFR TKIs treatment and reached the drug resistant state. There were also 16 treatment naïve patients in our study, who were recruited for detecting the primary EGFR T790M mutation.
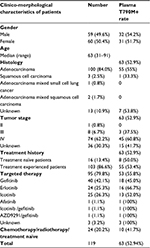  | Table 1 Clinico-morphological characteristics of patients |
Plasma EGFR T790M detection results by 3D Digital PCR
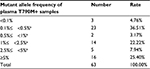
We performed a 3D Digital PCR analysis of 119 plasma ctDNA samples, to detect T790M mutation alleles from NSCLC patients. The percentage of T790M-mutant allele frequency in the 119 plasma samples was shown in Table 1, and the ratios of mutant allele frequency were summarized in Figure 1 and Table 2. The overall plasma EGFR T790M mutation rate in Chinese NSCLC patients was 52.94%, notable, eight of the 16 treatment naïve patients were found to be plasma T790M+. No statistical difference was found between the pretreatment T790M mutation and clinical parameters such as gender, histologic type, or tumor stage. Although there is a tendency that patients treated with erlotinib are more likely to acquire resistance of T790M mutation compared with gefitinib or icotinib, no statistical difference was found.
  | Figure 1 Detection of plasma EGFR T790M mutation alleles in NSCLC patient samples. Abbreviation: NSCLC, non-small cell lung cancer. |
Among the 63 positive cases, three samples (4.76%) presented at a low ratio (<0.1%) that could be detected with the 3D Digital PCR system. The median mutant allele frequency of plasma T790M was 1.09%. A total of 19 samples (30.16%) presented at a median ratio (<5%), and 16 samples (25.4%) presented at a high ratio (>5%) in this study.
Ultra-low plasma EGFR T790M detected by 3D digital PCR
To demonstrate the sensitivity of our 3D Digital PCR assay to detect plasma EGFR T790M, we compared the result of ctDNA extracted from one plasma sample detected by our assay with the CFDA-approved ADx-ARMS EGFR mutation kit. Limited by the amount of 10 mL extracted from a patient’s blood ctDNA, we selected 17 samples which had T790M frequency <2.5% and eight samples which were T790M-, to do ARMS-PCR single blind verification. We used McNemar’s test to analyze whether the results between 3D Digital PCR and ARMS-PCR were consistent, the results showed that: P=0.0002, indicating that the results of the two methods were inconsistent. Then we analyzed the consistency of T790M+ samples and T790M- samples between the two methods separately using Fisher’s exact test, the results showed that: P=0.0001, indicating that those were inconsistent, and the number of 3D Digital PCR T790M+ samples detected was much more than ARMS-PCR, which demonstrated the sensitivity of our 3D Digital PCR assay.
Four samples were enough for verification by Illumina HiSeq sequencer as a third verification method. The general comparison results were shown in Tables 3 and 4; we also enumerate the test results of ten of the samples using the three platforms in Table 5. Among the selected 17 positive cases, ten samples presented at a ratio of <0.5%, and seven samples presented at a ratio of <2.5%. However, only three samples were identified as positive for the EGFR T790M mutation by ARMS-PCR, namely, P6 (mutant allele frequency of T790M detected by 3D Digital PCR was 1.09%), P7 (mutant allele frequency of T790M detected by 3D Digital PCR was 2.09%), and P8 (mutant allele frequency of T790M detected by 3D Digital PCR was 2.21%). It is worth mentioning that sample P9 was identified as T790M- by ARMS-PCR, however, mutant allele T790M was detected by 3D Digital PCR at a frequency of 2.05%, suggesting that our 3D Digital PCR assay is more sensitive for detecting plasma T790M mutation. Samples P1–P4 was identified as T790M+ by both NGS and 3D Digital PCR, typically P1–P3 presented at a low ratio (<0.5%), suggesting the specificity of our 3D Digital PCR assay.
  | Table 3 Summary of 25 samples of T790M genotyping by 3D Digital PCR |
  | Table 4 Comparison results between 3D digital PCR and ARMS-PCR of the 25 samples |
  | Table 5 Triple platform detection results of ten samples |
Discussion
In the treatment process of EGFR-TKIs, most patients will eventually become resistant to EGFR-TKIs therapy. As we know, T790M mutation of EGFR gene is the most important mechanism of EGFR-TKIs drug resistance. More than 50% EGFR-TKIs drug resistance is caused by T790M mutation.17,37 It is reported that T790M mutation in peripheral blood occurred earlier than the disease progression.38 It is beneficial in terms of clinical decisions for patients, if T790M mutation is detected early. In addition, requiring additional biopsy tissue causes difficulties for those with advanced or recurring lung cancer. Because of tumor heterogeneity and the fact that biopsy is usually done at single site, there may be homogeneity in the genetic information from patient plasma.39 Compared with tumor tissue, detecting T790M mutation in the plasma ctDNA of lung cancer patients will have great advantages.40 Detecting T790M mutation in plasma is already a focus of research. There are some studies which used different assays for detecting EGFR gene status in plasma, including DHPLC, ARMS-PCR, and ddPCR.41 3D Digital PCR system is a relatively new technology of ddPCR.42 It requires no external calibrators for measuring the absolute and relative copy numbers of target DNA. Rapid microfluidic analysis of thousands of droplets per sample makes ddPCR practical for routine use. 3D digital PCR extends the performance of existing TaqMan assays, enabling applications that benefit from higher sensitivity, precision, or absolute quantification. In our study, in the group of 119 plasma ctDNA samples from NSCLC patients, 63 (52.9%) were found to have T790M gene mutation by 3D digital PCR. Among the 63 positive cases, three samples (4.76%) presented at a low ratio (<0.1%) that could be detected with the 3D digital PCR system. The median plasma T790M mutation frequency is 1.09%. Our T790M mutation frequency data according to ctDNA is consistent with the frequency (more than 50%) according to tissue sampling, which was shown in previous studies of patients after disease progression and treatment with EGFR TKIs.43,44 Therefore, our results show that 3D Digital PCR system is a sensitive method to detect EGFR T790M status in plasma of advanced NSCLC patients.
Ishii et al detected T790M status in EGFR-TKI-resistant patients by digital PCR. The result showed the sensitivity was 81.8%, and the specificity was 85.7%. The consistency with the paired tissue sample detection was 83.3%.45 Thress et al used four platforms respectively for detecting EGFR mutations in plasma ctDNA.33 The platforms included two non-digital platforms (Cobas® and ARMS-PCR) and two digital platforms (BEAMing digital PCR and Droplet digital PCR). For the T790M mutation, the sensitivity and specificity was 73% and 67%, respectively, with the Cobas® EGFR Mutation Test, and 81% and 58%, respectively, with BEAMing PCR. The research finding suggested that the Cobas® EGFR Mutation Test and BEAMing PCR demonstrate a high sensitivity for T790M mutation detection. Zheng et al’s research showed that T790M ctDNA mutation in plasma was detected in 55 (47%) of the 117 patients, using the ddPCR method.46 The results demonstrated that the overall concordance rate of T790M testing between the paired tumor tissue and plasma was 88.00% (22/25). The sensitivity and specificity of plasma T790M testing by ddPCR assay was 81.25% (13/16) and 100.00% (9/9), respectively. Therefore, according to the research findings, ddPCR is a potential method with higher precision, sensitivity, and accuracy for detecting EGFR gene T790M mutation in plasma compared with ARMS assay. In our study, we compared the 3D Digital PCR with the ARMS-PCR method for detecting T790M mutation. Our result showed only three plasma samples were identified as positive for the EGFR T790M mutation by ARMS-PCR in 17 T790M mutation positive cases detecting by 3D Digital PCR. These 17 positive samples included ten samples which presented at a ratio of <0.5%, and seven samples that presented at a ratio of <2.5%. Meanwhile four positive plasma samples were verified by NGS platform. Our data illustrated that 3D Digital PCR assay was more sensitive and specific than ARMS-PCR for detecting T790M mutation in plasma samples.
Regarding limitations of our study, firstly, we collected 8–10 mL peripheral blood from every patient according to their clinical statuses. Plasma ctDNA levels vary greatly among different patients. Because of the limitation of plasma sample volume, we did not compare all the samples with 3D Digital PCR and ARMS assay. Meanwhile, only four plasma samples were enough to be validated by NGS method. Secondly, we did not obtain corresponding tumor tissues to detect the T790M status because it is difficult to obtain biopsies from advanced NSCLC patients. So, we could not compare T790M status in plasma ctDNA with tumor tissues.
Conclusion
We evaluated a novel 3D Digital PCR assay for detecting EGFR gene T790M mutations in plasma samples of NSCLC patients. Our study indicated the great advantages of 3D Digital PCR regarding EGFR T790M mutation after resistance to EGFR-TKIs. 3D Digital PCR technology has more sensitivity and specificity than traditional PCR methods. 3D Digital PCR is a suitable method for plasma ctDNA testing in NSCLC patients with EGFR-TKI resistance.
Acknowledgment
We would like to thank Zhongwu Li, Lixin Zhou, and Yanhua Bai for assistance with the preparation of blood samples and Shenyi Lian for valuable discussion.
Disclosure
The authors report no conflicts of interest in this work.
References
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Hong QY, Wu GM, Qian GS, et al. Prevention and management of lung cancer in China. Cancer. 2015;121 Suppl 17:3080–3088. | ||
Reck M, Popat S, Reinmuth N, De Ruysscher D, Kerr KM, Peters S; ESMO Guidelines Working Group. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25 Suppl 3:iii27–iii39. | ||
Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546. | ||
Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. | ||
Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16(9):e447–459. | ||
Tan DS, Mok TS, Rebbeck TR. Cancer genomics: diversity and disparity across ethnicity and geography. J Clin Oncol. 2016;34(1):91–101. | ||
Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–742. | ||
Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. | ||
Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–128. | ||
Kim ES, Hirsh V, Mok T, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet. 2008;372(9652):1809–1818. | ||
Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327–3334. | ||
Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15(2):213–222. | ||
Shi Y, Zhang L, Liu X, et al. Icotinib versus gefitinib in previously treated advanced non-small-cell lung cancer (ICOGEN): a randomised, double-blind phase 3 non-inferiority trial. Lancet Oncol. 2013;14(10):953–961. | ||
Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31(8):1070–1080. | ||
Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105(6):2070–2075. | ||
Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. | ||
Lin Y, Wang X, Jin H. EGFR-TKI resistance in NSCLC patients: mechanisms and strategies. Am J Cancer Res. 2014;4(5):411–435. | ||
Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19(11):1389–1400. | ||
Xu X, Mao L, Xu W, et al. AC0010, an irreversible EGFR inhibitor selectively targeting mutated EGFR and overcoming T790M-induced resistance in animal models and lung cancer patients. Mol Cancer Ther. 2016;15(11):2586–2597. | ||
Kim ES. Olmutinib: first global approval. Drugs. 2016;76(11):1153–1157. | ||
Sequist LV, Soria JC, Gadgeel SM, et al. First-in-human evaluation of CO-1686, an irreversible, highly selective tyrosine kinase inhibitor of mutations of EGFR (activating and T790M). J Clin Oncol. 2014;32:15 Suppl:8010. | ||
Greig SL. Osimertinib: first global approval. Drugs. 2016;76(2):263–273. | ||
Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376(7):629–640. | ||
Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. | ||
Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. | ||
Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. | ||
Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–540. | ||
Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013; 10(8):472–484. | ||
Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. | ||
Holdhoff M, Schmidt K, Donehower R, Diaz LA Jr. Analysis of circulating tumor DNA to confirm somatic KRAS mutations. J Natl Cancer Inst. 2009;101(18):1284–1285. | ||
Xu T, Kang X, You X, et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor DNA from non-small cell lung carcinoma patient plasma. Theranostics. 2017;7(6):1437–1446. | ||
Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–515. | ||
Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3375–3382. | ||
Gu J, Zang W, Liu B, et al. P2.03b-098 comparison of digital PCR, ion proton with ARMS-PCR in tumor tissue and plasma of NSCLC patients. 2017;12(1):S996. | ||
Yao Y, Liu J, Li L, et al. Detection of circulating tumor DNA in patients with advanced non-small cell lung cancer. Oncotarget. 2017;8(2):2130–2140. | ||
Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. | ||
Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer. 2014;120(24):3896–3901. | ||
Ichihara E, Lovly CM. Shades of T790M: intratumor heterogeneity in EGFR-mutant lung cancer. Cancer Discov. 2015;5(7):694–696. | ||
Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a Phase I study of rociletinib (CO-1686). Clin Cancer Res. 2016;22(10):2386–2395. | ||
Ke EE, Wu YL. EGFR as a pharmacological target in EGFR-mutant non-small-cell lung cancer: where do we stand now. Trends Pharmacol Sci. 2016;37(11):887–903. | ||
Conte D, Verri C, Borzi C, et al. Novel method to detect microRNAs using chip-based QuantStudio 3D digital PCR. BMC Genomics. 2015;16:849. | ||
Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. | ||
Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. | ||
Ishii H, Azuma K, Sakai K, et al. Digital PCR analysis of plasma cell-free DNA for non-invasive detection of drug resistance mechanisms in EGFR mutant NSCLC: correlation with paired tumor samples. Oncotarget. 2015;6(31):30850–30858. | ||
Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.