Back to Archived Journals » Oncolytic Virotherapy » Volume 8
Virus–Receptor Interactions: Structural Insights For Oncolytic Virus Development
Authors Jayawardena N, Burga LN, Poirier JT , Bostina M
Received 6 June 2019
Accepted for publication 2 October 2019
Published 29 October 2019 Volume 2019:8 Pages 39—56
DOI https://doi.org/10.2147/OV.S218494
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chae-Ok Yun
Nadishka Jayawardena,1 Laura N Burga,1 John T Poirier,2 Mihnea Bostina1,3
1Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand; 2Perlmutter Cancer Center, NYU Langone Health, New York, NY, USA; 3Otago Micro and Nano Imaging, University of Otago, Dunedin, New Zealand
Correspondence: Mihnea Bostina
Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand
Tel +64 22 44 5583 Email [email protected]
Abstract: Recent advancements in oncolytic virotherapy commend a special attention to developing new strategies for targeting cancer cells with oncolytic viruses (OVs). Modifications of the viral envelope or coat proteins serve as a logical mean of repurposing viruses for cancer treatment. In this review, we discuss how detailed structural knowledge of the interactions between OVs and their natural receptors provide valuable insights into tumor specificity of some viruses and re-targeting of alternate receptors for broad tumor tropism or improved tumor selectivity.
Keywords: oncolytic viruses, virus–receptor interaction, virus entry
Introduction
Oncolytic virotherapy is a dynamic field of cancer treatment with over 70 clinical trials registered to date.1 The majority of oncolytic viruses (OVs) are used in their native, replication-competent form to cause a direct oncolysis of tumors. For instance, coxsackievirus, parvovirus, Newcastle disease virus, measles virus, vaccinia virus and Seneca Valley virus have been used in clinical trials in their native forms.2–6 On the other hand, human pathogenic viruses such as herpes simplex virus-1, poliovirus and adenovirus have been genetically modified to limit their replication to tumor sites and to reduce their virulence in normal tissues.7–9 In addition to the direct oncolysis, OVs can kill cancer cells via several indirect mechanisms: the activation of immunologic pathways and antiangiogenesis.10,11 En route to reaching cancer cells, OVs must overcome a range of complex physical and chemical barriers to finally interact with specific cellular receptors.12 Perhaps the most exhaustive obstacle in systemic delivery of OVs is the neutralization of viruses by pre-existing antibodies or triggered anti-viral immune response.13 One way to bypass the host immunity is to mask/manipulate viral surface proteins to avoid recognition by neutralizing antibodies.14,15 However, eliminating antibody recognition does not guarantee a successful infection of tumors with OVs as the cellular uptake will ultimately be dependent on virus binding to the cellular receptors. Expression of virus cellular receptors in cancers varies depending on tumor type as well as among different patients with the same type of cancer.16 In such cases, OVs need to be modified to re-target the cancer via alternative receptors. Thus, the manipulation of OV surface proteins to either circumvent anti-viral immune response or to exploit different receptors requires in-depth knowledge of how they interact with their cellular receptors at a structural level. In this review, we discuss the interactions between clinically evaluated OVs and their cellular receptors and how they have been modified to target cancers.
Oncolytic Viruses And Cancer Tropism
Herpes Simplex Virus
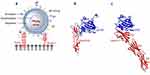
Herpes simplex virus 1 and 2 (HSV-1 and HSV-2) belong to the family of Herpesviridae, genus Simplexvirus.17 HSV virion has a complex architecture characterized by a dsDNA genome, an icosahedral capsid (nucleocapsid), an amorphous layer of protein (tegument) and an envelope (Figure 1A).18 Both HSV-1 and -2 are genetically stable and considered to be the most serious human pathogens in their family. HSV-1 was shown to be associated with encephalitis and orofacial herpes infections, whereas HSV-2 mostly causes genital infections.19 The remarkable pathogenicity of HSV is attributed to its ability to establish latent infections in sensory neurons, thus providing a logical reason to manipulate these strains for therapeutic applications.
T-VEC is a genetically modified strain of HSV-1 and represents a major breakthrough in immunotherapy being the first and only US FDA approved oncolytic virus to date.20–23 T-VEC is presently used as intralesional injections to treat non-resectable melanoma with many ongoing Phase I/II clinical trials showing the possibility of using the virus in conjunction with other treatments such as immune checkpoint inhibitors.24,25 Furthermore, two other strains of HSV-1, G207 (Infected cell protein (ICP) 34.5 and ribonucleotide reductase mutated) and NV1020 (ICP34.5 mutated) have completed Phase I/II clinical trials in malignant brain tumors and in colorectal cancer liver metastasis, showing partial clinical responses and stabilization of metastasis, respectively.26,27
While modifications to the T-VEC genome are aimed at reducing the neurotoxicity of the wild-type strain and stimulating a strong immune response in tumor site, expression levels of cellular receptors and their interactions with HSV still play a vital role in virus entry into tumor cells. HSV utilizes four viral glycoproteins, gB, gD, gH and gL (Figure 1A), expressed on the outer envelope to establish interactions with various cell surface receptors and to facilitate cell entry.28,29 In order to initiate HSV cell entry, at least three different classes of cell surface receptors should interact with the respective glycoproteins.29,30 Current molecular and structural biology literature identifies three steps in penetrating host cells: 1) gB attachment to heparan sulfate proteoglycans (HSPG)244, 2) gD binding to nectin-1,31,32 herpes virus entry mediator (HVEM),33 or 3-O-sulfated heparan sulfate, and 3) gB binding to paired Ig-like type 2α (PILRα),34 nonmuscle myosin IIA (NMHC-IIA) or myelin-associated glycoprotein (MAG) and initiation of envelope fusion with plasma membrane.29 Upon the envelope fusion with host-cell membrane, HSV nucleocapsid is translocated to the nuclear pore through which viral DNA is released into the nucleus.35 Evidence from various clinical studies points toward a direct relationship between the expression of HSV receptors in tumors, cancer progression and prognosis. For instance, herpesvirus entry mediator (HVEM), a member of the tumor necrosis factor (TNF) superfamily, has been shown to play a role in activating inhibitor signaling in T-cells upon binding to BTLA ligand (B-lymphocyte and T-lymphocyte attenuator).36 Increased expression of HVEM has been reported in hepatocellular carcinoma,37 gastric cancer38 and melanoma.36 Structural evidence for interactions between HVEM and HSV gD protein arises from a crystal structure of gD ectodomain truncated at residues 285 (gD285) bound to the ectodomain of HVEM (Figure 1B).33 In both HSV-1 and -2, gD is structurally unique in comparison to other members of the family due to diverging N-terminal hairpins.39 The interface between gD285 and HVEM is comprised of interactions between short, N-terminal hairpin (1–37) that extends towards the V-like immunoglobin core of gD to establish interactions with two cysteine-repeat-domains (CRDs) of HVEM. C-termini of gD and HVEM are arranged in opposite directions, presumably anchored to viral and cellular membranes, respectively. The observation that only a small segment of gD protein is involved in HVEM binding suggests the possibility of manipulating gD protein to redirect HSV to a different receptor, such as nectin-1 or 3-O-sulfated heparan sulfate, depending on their expression levels in cancers.14 Nectin-1 is another cell surface receptor that binds gD of HSV.32 Nectin-1 belongs to the family of nectin or nectin-like receptors that play an important role in cell adhesion.40 Results from various in vitro and clinical studies have identified increased expression of nectin-1 and nectin-2 in cancers such as breast cancer,41 highly migratory and invasive carcinoma,42 squamous cell carcinoma43 and colorectal cancer.44 In such instances, nectin-1 serves as an excellent predictor of HSV oncolytic sensitivity. Interactions between gD and nectin-1 have been characterized by crystallization of truncated forms of the gD ectodomain (gD285, truncated residues 1–285) complexed with nectin-1 (Figure 1C).31 The crystal structure identifies both N- and C-termini and a residue located in Ig core interacting with the first Ig domain of nectin-1 at 1:1 stoichiometry, resembling an interaction pattern similar to nectin-1 homodimers. Interestingly, interactions in gD285-nectin 1 interface are similar to those observed in nectin-1 homodimer interface and distant from gD285-HVEM interface due to the absence of N-terminal hairpin. From a physiological point of view, gD binding to nectin-1 can abolish nectin-1 dimerization, eventually affecting cell–cell adhesion.31 Therefore, modified HSV strains could have an additional mechanism of hampering tumor progression apart from triggering anti-tumor immunity.
Because of the wide expression of nectin-1 in human cells,45 targeting nectin-1 expressing tumors with HSV-1 could be problematic in the case of systemic immunotherapy. Such off-target effects can be minimized either by developing HSV mutants capable of escaping nectin-1 while still retaining its ability to bind HVEM, or by identifying potential bi-soluble adapters for targeting cognate tumor receptors.46 First evidence for latter strategy comes from targeting of epidermal growth factor receptor (EGFR) expressing cells with a HSV variant modified with P-V528LH adapter consisting of gD ectodomain binding region of nectin-1 fused to an EGFR-specific monoclonal antibody.46
Vaccinia Virus
Vaccinia virus (VV) is a large, enveloped dsDNA virus (~191 kbp) from the genus Orthopoxvirus of the Poxviridae family.47 The natural host and origin of VV are not known.48 Characteristic to VV is its replication strategy which takes place in cytoplasmic viral factories of infected cells.49 The genome of VV encodes more than 200 proteins, of which approximately 20 are envelope proteins.50 During the life cycle of VV, three distinct particle types are produced; (1) intracellular mature virions (IMV), (2) wrapped virions (WV) and (3) extracellular enveloped virions (EEV).50 Mature virions (MV) are stable under virus purification conditions, remaining the most extensively studied form of the virus. By contrast to other dsDNA viruses, IMV has a complex, asymmetric structure that consists of a nucleoprotein core surrounded by a single lipoprotein membrane.51
Since its use in eradicating smallpox,52 VV has played a seminal role as recombinant vectors in gene therapy.53 Both wild-type and recombinant strains of VV have been of particular interest in oncovirotherapy.54 As an oncolytic agent, VV has several advantages such as the ability to incorporate a large amount of foreign DNA, fast and efficient replication and safety.55 Moreover, VV displayed natural cancer tropism, selectively targeting tumors after systemic administration.54 Clinical trials on VV thus far have employed a potent, yet safe form of VV (JX-594), which encodes granulocyte-macrophage colony-stimulating factor as an immunomodulator.55,56,57
Vaccinia virus MVs entry into host cells is either mediated by fusion of MV membrane with the plasma membrane at neutral pH or through receptor-mediated endocytosis under acidic conditions.58,59 Nonetheless, no receptors have been unequivocally identified. Glycosaminoglycans (GAGs), highly polyanionic compounds present on the surface of stromal tumor cells, have been suggested as putative receptors facilitating VV entry.59,60 VV membrane proteins A27L and H3L are essential for fusion of viral membrane with cell membrane.61,62 Positively charged amino-terminal of A27L can also act as a site for binding of heparan sulfate (HS).63 The involvement of additional GAGs such as chondroitin sulfate (CS) in binding the VV surface protein D8L has been shown, but subsequent studies eliminated the essentiality of these receptors.59,64,65 To date, an exact mechanism behind VV-induced oncolysis is unknown. Whether the anti-tumor efficacy is receptor-mediated or attributed to tumor vasculature66 or whether overexpression of ribonucleotide reductase is essential for viral replication67 still remains an open question.
Rhabdoviruses
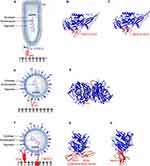
Members of Rhabdoviridae family are enveloped, negative-sense single-stranded (ss) RNA viruses with a 11–15 kb linear genome encoding five proteins: glycoprotein (G), matrix protein (M), phosphoprotein (P), polymerase (L) and nucleoprotein (NP).68 Rhabdoviruses (RhVs) virions are about 180 nm long and 75 nm wide and have a rod- or bullet-shaped geometry. The G proteindecorating the envelope is involved in receptor binding, whereas NPs are associated with RNA (NP-RNA). Together with L and P, NP-RNA complex forms a ribonucleoprotein particle, which makes contact with M proteins beneath the envelope (Figure 2A).
RhVs have a broad and diverse host specificity with Lyssavirus and Vesiculovirus genera, infecting animals and the remaining RhVs infecting plants.69 RhVs present several advantages that recommend them for development as oncolytic agents. RhV infections are relatively rare, therefore there is no pre-existing immunity. Additionally, they do not show genetic reassortment, integration in the host genome or malignant transformation due to cytoplasmic replication and have a relative ease of large-scale virus production in a broad range of cell lines. Several RhVs have been investigated for their oncolytic properties.70,71
Vesicular stomatitis virus (VSV) is a vesiculovirus that infects cattle, horses, pigs, and other mammals. VSV infections are usually asymptomatic in human and non-lethal in animals, with mild flu-like symptoms.70 VSV exhibits a robust infectivity and broad tropism to tumors, attributed to the defective interferon (IFN) responses in tumor cells.72 Entry of VSV into tumor cells is initiated by the interactions between its coat protein VSV-G (Figure 2A) and highly ubiquitous cellular receptor, low-density lipoprotein receptor (LDLR).73 LDLR is a transmembrane receptor whose functions include cell-signaling, endocytosis and trafficking of cellular proteins. The most abundantly expressed form of LDLR in solid tumors is LDLR1, shown to be linked to low patient survival rate.74 The ligand binding domain of LDLR is comprised of cysteine-rich repeats conserved among other members of LDLR family,75 therefore presenting alternative entry points for VSV. Crystal structures of VSV-G in complex with two different cysteine-rich domains, CRD2 (Figure 2B) and CRD3 (Figure 2C) of LDLR demonstrate that both binding sites on VSV-G are identical.76 VSV-G-LDLR complex is internalized into host cells through a clathrin-mediated endocytosis.77,78 In the case of recombinant VSVΔM51 encoding reovirus fusion-associated small transmembrane (FAST) protein, this mechanism extends from the virus-cell fusion to cell-cell fusion.79 The process repeats, expanding to un-infected cells and could lead to large multinucleated giant cells (syncytia).80 In another study, the use of VSV-G substituted with lymphocytic choriomeningitis virus glycoprotein (LCMV-GP) has shown minimal neural toxicity and potent anti-tumor effect in mice brain tumor models.81 LMCV-GP may bind differentially glycosylated α-dystroglycan (αDG) in brain tumors with high-affinity82,83 despite the lower expression levels of αDG in human glioblastoma.84
Maraba virus is another vesiculovirus which binds LDLR and has the capacity to infect a broad range of human cancers.85,86 In order to specifically target cancer cells and to enhance replication efficacy, two mutations were introduced: L123W and Q242R in M and G proteins, respectively.87,88 Maraba virus strain MG1 expressing human melanoma-associated antigen-A3 (MAGE-A3) and an adenoviral vector (Ad) expressing the same antigen have been developed as an oncolytic vaccine strategy with high immune priming efficiency.89
Newcastle Disease Virus
Newcastle disease virus (NDV) is a ssRNA virus in the genus Avulavirus of Paramyxoviridae family.90 The enveloped NDV capsid harbours a non-segmented negative-sense ssRNA that codes for six proteins (Figure 2D). Nucleoprotein (NP), phosphoprotein (P) and RNA dependent RNA polymerase (RdRP) bind the RNA genome to form the nucleocapsid.90 Other NDV proteins include matrix protein (M), which forms the inner layer of virus envelope, hemagglutinin-neuraminidase (HN) and fusion protein (FP), involved in receptor binding and entry, respectively.91
Numerous in vitro studies have shown that NDV is non-pathogenic to humans and elicits anti-tumor effects without any genetic modifications or limitations in delivery methods.92–94 MTH68/H, PV701 and NDV-HUJ are three attenuated strains of NDV with highly efficient intratumoral replication, tumor cell lysis and immunostimulation, currently in Phase I/II clinical trials.4,95 In addition, NDV-HUJ strain is able to bypass the effect of an anti-apoptotic protein Livin.4
NDV binds tumor cells via interactions between HN and cell surface sialic acids (SA) receptors.96 SA is a derivative of neuraminic acid overexpressed in multiple cancers145–147 and was shown to be associated with the metastasis of breast cancer.97 HN has a dual function: to recognize the cell surface receptors, and subsequently to promote the fusion activity of the F protein and to cleave off the sialic acid from progeny virus particles.98 HN is composed of a long stalk connected to a globular head that consists of a six-bladed β-sheet propeller.99 Two sites have been identified on HN dimers that form interactions with sialic acid (Figure 2E). The first binding site is involved in mediating neuraminidase activity, receptor binding and promoting the fusion activity of F protein.100 The second binding site is located at the membrane-HN distal region interface, which aids in tethering the virus in close proximity to the host membrane during fusion.101,102
Measles Virus
Measles virus (MV) is an enveloped, spherical-shaped, negative-sense ssRNA virus (Figure 2F) from the genus Morbillivirus of the Paramyxoviridae family.103 Due to its highly contagious nature, MV remains a major human health concern worldwide, causing approximately 150,000 deaths annually.104 Similar to RhVs, the non-segmented RNA genome (15–16 kb in size) of MV encodes five structural proteins: glycoprotein (G), matrix protein (M), phosphoprotein (P), large protein (L) and nucleoprotein (NP).103,105 On the MV envelope there are two types of glycoproteins characteristic to paramyxoviruses: 1) hemagglutinin106 and 2) fusion protein,107 responsible for cell receptor attachment and fusion, respectively.
Live-attenuated MV vaccine strains can be used as oncolytic agents to target different receptors overexpressed in tumors.3 Measles virus hemagglutinin (H) binds CD46,108 signaling lymphocyte activation molecule (SLAM)109 or nectin-4 in epithelial cells.110 Overexpression of CD46 and nectin-4 receptors has been identified as a strategy to preferentially target cancers with MV.111,112 In addition, SLAM expressed on activated B and T-lymphocytes, monocytes, and dendritic cells has been reported to be the main entry port for wild-type MV.113
CD46 structure is comprised of a C-terminal domain, a transmembrane domain, a short region with unknown function and four modules of short consensus repeats (SCR) 1-4 at the N terminus.143,114 The crystal structure of dimeric H-CD46 identifies interactions between MV-H and SCR1, SCR1-SCR2 interface and SCR2 of CD46 (Figure 2G). CD46 SCR1-2 is pivotal to capsid binding in adenoviruses,115 discussed later in this review. However, the binding sites of MV-H for CD46 and SLAM overlap,116 supporting the need to develop strains that can preferentially bind CD46. Two amino acid substitutions, N481Y and S546G in MV-H protein, have been shown to arm MV strains to efficiently use CD46 as an entry receptor in CD46+ cells.117 In another study, structural characterization of MV-H-nectin-4 complex revealed that the amino-terminal of nectin-4 binds β4-β5 groove of MV-H (Figure 2H).110 This study identified a hydrophobic pocket located in the groove suggested to be involved in binding all three receptors for MV, with different residues involved for different receptors.110
Similar to RhVs, MV exerts its oncolytic activity by a sequence of virus-cell fusion through H protein, cell-cell fusion through F protein and subsequent apoptosis.118 The Edmonston vaccine strain of MV (MV-Edm) has been modified for non-invasive imaging of MV activity in tumors by introducing either sodium iodide symporter (NIS), the β subunit of human chorionic gonadotropin (βhCG) or human carcinoembryonic antigen (CEA) into the MV genome. The MV-CEA strain has been tested in Phase I clinical trials in patients with platinum-resistant ovarian cancer, with evidence pointing towards the recruitment of anti-tumor effector T-cells to establish an anti-tumor immunity.119,120 Selective tumor tropism of MV was further validated in a Phase I clinical trial in myeloma, where systemically administered MV-NIS showed replication within tumors.121 Alternative to live-attenuated vaccines, recombinant, replication-competent MV could be developed to re-target different surface receptors expressed on tumor cells. This requires the mutation of SLAM and CD46 binding sites, thus resulting in a double ablated chimeric H protein to prevent the binding of MV to normal cells expressing SLAM and CD46.122 Mutations at Y481 and R533 on MV-H and subsequent incorporation of single-chain antibodies directed against cognate receptors such as EGFR has shown to elicit oncolysis of EGFR-positive tumor models in mice.123,124
Adenovirus
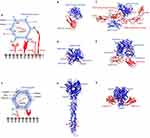
Human adenoviruses (HAdVs) belong to the family of Adenoviridae, genus Mastadenovirus and are divided into seven different species from HAdV-A to -G.125 They are non-enveloped viruses with an icosahedral capsid protecting a dsDNA genome of 26–46 kbp (Figure 3A). The icosahedral capsid is comprised of 252 capsomeres, consisting of 240 hexon trimers and 60 penton bases (PB).38,126 Attached to PB plates are trimers of fiber molecules, which utilize the conserved N-terminal (residues 1–20) to bind the PB and the C-terminal knob to bind cellular receptors. Collectively, hexon trimers, PBs, and fiber molecules are known as the major capsid proteins. In addition, 240 copies of the minor protein IX, and several copies of the minor proteins IIIa, VI and VII are located on the capsid exterior and interior, respectively. All the HAdV strains can cause gastrointestinal infections, with some subtypes being reported to cause respiratory, urinary tract infections and keratoconjunctivitis.127 HAdV is also responsible for viral-induced tumors in mice, with subtype A showing the highest oncogenicity, subtype B being weakly oncogenic,128,129 while C, E and F are known to be non-oncogenic.127
Adenoviruses are one of the most extensively studied viral vectors due to ease of genome manipulation. In addition, HAdVs provide several distinct advantages such as inherently potent lytic activity and feasibility of manufacturing high viral titers.130 Numerous HAdV strains have been genetically engineered (ONYX-015 and DNX-2401) to reduce infection in normal tissues and to selectively target tumors.131,132
HAdV entry into host cells is a two-step mechanism, which involves the initial attachment of the viral fibers to cell surface receptors, followed by interactions with other capsid proteins and internalization receptors.133 Upon virus internalization by endocytosis, the capsid escapes into the cytosol through lysis of endosomal membrane and is subsequently trafficked to the nuclear envelope along microtubules, where the viral genome enters the host nucleus via nuclear pores.134 Most of the HAdVs and other AdV subtypes except HAdV B use the coxsackievirus and adenovirus receptor (CAR) for cellular attachment.135 CAR is a type I transmembrane glycoprotein that belongs to the immunoglobulin (Ig) superfamily. It contains a cytoplasmic C-terminal, a hydrophobic transmembrane domain and two extracellular Ig-like domains, D1 and D2.136 CAR D1 domain alone is sufficient to establish interactions with HAdV fiber knob (Figure 3B).137,138 However, the variable expression of CAR in cancers is a significant challenge for HAdV oncovirotherapy.139 Low expression of CAR was reported for gastric, colon, and prostate cancer cell lines under hypoxic conditions.140 In addition, CAR expression is downregulated in cancer cells treated with chemotherapy or radiation, which poses an issue when using HAdVs in combination therapies.141
The majority of subtype B HAdVs and some of subtype D (AdV37) have been shown to exploit CD46, a type I transmembrane protein overexpressed in tumors, as the attachment receptor.142,143 Crystallographic studies showed that AdV11 trimeric fibers form a compact knob that interacts with the SCR1-2 regions of three CD46 molecules (Figure 3C).115 Complementary or not to CD46, sialic acid has been shown to interact with the top region of AdV37 knob trimer. This has been further confirmed by structural studies on AdV52, which utilizes its long fibers to bind CAR and short fibers to bind polysialic acid (Figure 3D).144,145,146,147 CD80 and CD86 are another two members from the Ig superfamily that play a key role in subtype B AdV3 entry. Both CD80 and CD86 are expressed in dendritic cells, thus targeting these receptors by AdV can elicit a strong immune response via T-cell activation.148,149,150
Recent studies have identified desmoglein 2 (DSG2) as a new receptor for HAdV3, HAdV7, HAdV11 and HAdV14 strains of subtype B.151 DSG2 is a type 1 transmembrane glycoprotein present in epithelial cells that plays an essential role in cell-cell adhesion.152,153 The extracellular domain of DSG2 is comprised of four cadherin domains, EC1-EC4, with EC2 and EC3 accounting for the region that binds trimeric fiber knob of HAdV3.154 DSG2 is overexpressed in a range of epithelial cancers, acting as a marker for targeting such cancers with AdV.155,156,157,158 A cryo-EM study showed that DSG2 EC2-EC3 fragment binds the top of the trimeric HAdV3 in 2:1 or 1:1 stoichiometry (Figure 3E).159 EC2 and EC3 establish interactions with the loop regions of monomers 1 and 2, respectively, while the third HAdV monomer of the knob is not engaged. Furthermore, mutagenesis experiments identified D261 as an essential knob residue required for DSG2 binding.159
Endocytosis of the HAdV-CAR complex is mediated by the interactions between internalization receptors, integrins and five-fold capsid vertices.160 Structural information is available on entry receptors αvβ3 integrins bound to adenovirus, which shows the requirement of Arg-Gly-Asp (RGD) moiety on the penton base to interact simultaneously with several integrins in different orientations to facilitate integrin clustering and subsequent viral entry into host cells via endocytosis.161,162 However, mutation of RGD sequence was associated with only a reduced viral infection but not complete abolishment.163 Though no plausible mechanisms have been proposed for an integrin-independent entry pathway of AdV, there is evidence for compensation for loss of penton-integrin interactions through recruitment of fiber receptors.163,164 In the absence of sufficient levels of CAR for a successful infection, re-targeting of integrin receptors by incorporation of an RGD moiety in the fiber knob of AdV5 has shown to be efficient in promoting infection of ovarian tumor cells.165
Reovirus
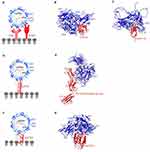
Reoviridae is a family of non-enveloped, dsDNA viruses with an icosahedral capsid structure (~85 nm in diameter) composed of a large outer layer, and a smaller inner layer (Figure 3F). Reovirus (RV) dsDNA is structured into 12 segments, categorized into three size-dependent groups: large, medium and small.166,167 The outer shell of the capsid and at the vertices of the virion are formed by heterodimers of µ1 and σ3 proteins, while pentamers of λ2 protein form a channel connecting to trimers of attachment protein σ1.168
RV requires interactions with junctional adhesion molecule-A (JAM-A)169 and cell surface monosaccharides such as sialic acid170 to penetrate the host cell. JAM-A expression has been proposed to be linked to tumor cell proliferation and progression, whereas in some cases an inverse relationship was observed.171 The first step of the RV binding to host cells involves a low-affinity interaction of the lower part (stalk) of σ1 protein with cell surface sialic acid (Figure 3G). This process facilitates the anchoring of RV capsid in close proximity to host-cell membrane in order to initiate interactions with a secondary receptor. High-affinity interactions between JAM-A D1 domain and the head domain of σ1 protein (σ1H)169 serve as the second step in RV host-cell attachment (Figure 3H).
Reolysin, a wild-type, non-pathogenic, serotype 3 RV, has been widely investigated in preclinical and clinical settings.172 Phase I and II clinical trials of advanced solid tumors and recurrent gliomas,173–175 and combination therapy with paclitaxel/carboplatin or docetaxel176,177 showed Reolysin to be safe and effective.
Parvovirus
Human parvovirus (HPV) is a single-stranded DNA virus in the Parvoviridae family, associated with a wide variety of diseases in humans.178 The genome of HPV is packaged inside an icosahedral capsid of ~280 Å in diameter. The capsid is composed of 60 structural subunits, in which major capsid protein VP2 is the primary protein (~95%) while the minor capsid protein VP1 is less abundant (~5%).179 Capsid proteins have an eight-stranded, antiparallel β-barrel “jelly roll” fold. Engineered and wild-type strains of HPV demonstrate a tumor-selective replication with excellent safety profiles. Oncolytic activity of HPV is attributed to the direct oncolysis as well as induced anti-tumor immunity.180
HPV B19 strain binds the erythrocyte P19 antigen expressed in erythroid progenitor cells.181 However, entry into host cells requires the involvement of α5β1 integrin co-receptor,182 known to be essential for tumor progression in certain cancers.183 Modifications of I367S and H373R in the dimple region of the capsid in rat parvovirus strain H-1PV have engineered the virus to re-target integrin receptors expressed in cancers.184 In another study, transferrin receptor 1 (TFR1) has been identified as the cellular receptor for canine parvovirus (CPV).185 TFR1 is a membrane glycoprotein linked to many diseases including cancers.185 However, TFR1 expression is variable across different cancers.186 The structure of CPV-TFR1 complex demonstrates an example for a receptor saturating only a few of the 60 equivalent binding sites on the capsid, resulting in an asymmetric interaction.185
Coxsackievirus
Coxsackievirus (CV) is a non-enveloped, positive-sense ssRNA virus (~7.4 kb) from the family of Picornaviridae, genus Enterovirus (Figure 4A). CV is a major human pathogen causing a number of diseases including myocarditis and meningoencephalitis.187 CV serotypes are categorized into two groups; (1) coxsackievirus A (CVA) and coxsackievirus B (CVB).188 CV RNA genomes code for four structural proteins VP1–VP4 that form an icosahedral capsid, and seven non-structural proteins.189,190 Characteristic to enteroviruses is the presence of four types of particles in their life cycle: mature virion, procapsid devoid of RNA, an expanded A-particle and an empty particle after RNA exit.191
Receptor binding in enteroviruses takes place in the “canyon”, a depression located at 5-fold axis of the capsid. The binding of the receptor displaces a fatty acid molecule called the “pocket factor” located in a hydrophobic pocket within VP1, below the canyon base. Loss of the pocket factor induces a series of conformational changes in capsid architecture, leading to capsid expansion and externalization of VP1 N-terminus as well as VP4 for membrane anchoring and subsequent RNA transfer.192 This mechanism holds true for most of the enteroviruses and has been well characterized for poliovirus (PV),193,194 enterovirus 71 (EV71)195 and CV196.
CVs utilize three different receptors for cellular entry. CAR acts as both attachment and entry receptor for CVB3.196 Cryo-EM reconstruction of CVB3 bound to full length human CAR has shown that the N-terminal region of CAR D1 domain contains the binding sites for CVB3 (Figure 4B).196 In the CVB3-CAR interface, A and G β strands of D1 domain form contacts with the north and south rims of CVB3 canyon. All the external CVB3 capsid proteins are involved in CAR binding with majority of the interactions localized to VP1. Of note is the moderately conserved nature of these receptor binding residues across six different CVB serotypes.197 VP2 residue N165 has been suggested to be critical in stabilizing the electrostatic interactions between the capsid and CAR.198 The distal end of CAR D1 domain is a shared site for CVB3 and adenovirus (as previously discussed) and their binding sites overlap on C β strand and FG loop.196 Additionally, the involvement of decay-accelerating factor (DAF) as an attachment receptor in the CVB3-RD strain has been demonstrated by another cryo-EM study,199 with one DAF molecule linking two adjacent protomers on the capsid exterior. The northern end of the VP2 puff (residues 129–180) in one protomer is linked to the south end of the puff of the adjacent protomer, and the bulk of the interactions are condensed between short consensus repeat (SCR) 2 and the north end of the puff. Unlike CAR D1 domain, DAF does not enter the canyon and thus, does not induce the conformational changes required for genome delivery into the host cell.200
On the other hand, CVA binds both DAF and intercellular adhesion molecule-1 (ICAM-1).201,202 DAF binding does not induce conformational changes and primarily acts as an attachment receptor,203 whereas ICAM-1 acts as an attachment/entry receptor for CVA. Overexpression of DAF and ICAM-1 has been reported in multiple cancers.204,205,206,207,208 ICAM-1 is a transmembrane immunoglobin with three structural components: extracellular N-terminus, transmembrane domain, and cytoplasmic C-terminus.209 Structural insights into CVA-ICAM-1 stem from several cryo-EM investigations (Figure 4C).201,202 Similar to other enteroviruses, the canyon of CVA24v binds ICAM-1 D1 domain at the quasi 3-fold axis of the capsid.202 The CVA24v-ICAM-1 interface is comprised of interactions established between the FG loop of ICAM-1 D1 domain and VP1. Finally, C and D β strands of ICAM-1 D1 domain interact with the VP1 GH loop, whereas the DE loop of D1 forms additional contact with VP2 in CVA24v. This study also provides insights into adapting CVA strains for a sialic acid binding as a secondary receptor by mutating residue 250 of VP2 to tyrosine. Sialic acid metabolism has been shown to be upregulated in metastatic cancers and acts as a receptor for other oncolytic viruses discussed here such as adenovirus, Newcastle disease virus and reovirus.
The therapeutic potential of CVA21 or CAVATAK has been investigated in various preclinical melanoma studies as monotherapy210 or in combination with doxorubicin.211 Furthermore, CAVATAK has completed a Phase I clinical trial in patients with advanced melanoma with promising safety and anti-tumor activity recorded.22 In vivo studies of non-small-cell lung cancer xenograft models treated with CVB3 demonstrated abscopal effect of this therapy, suggesting an enhancement of antitumor immunity.212
Poliovirus
Poliovirus (PV), a member of Enterovirus genus, family Picornaviridae, is the main causative agent of paralytic poliomyelitis.213 Three different PV serotypes can be differentiated according to their antigenic properties.214 Similar to CV, PV possesses a negative-sense ssRNA genome of 7.5 kb coding for seven non-structural proteins and four structural proteins, which constitute the icosahedral capsid (Figure 4D) as previously described for CV.215,216
Poliovirus entry into host cells is initiated by the interactions between poliovirus receptor CD155, and capsid canyon.217–220 PV undergoes the same conformational changes characteristic to other enteroviruses. A-particle formation,194,221 exit of RNA genome at a location on 2-fold axis193 and empty particle formation222 have been extensively characterized. CD155 is an onco-immunologic protein overexpressed in human cancers with a role in tumor cell invasion and migration.223 CD155 is a type I immunoglobulin-like transmembrane protein that contains three ectodomains D1–D3.218,224 CD155 expression is upregulated in carcinomas225–227 and less abundantly expressed in normal tissues with the exception of liver development or regeneration.228 In the PV-CD155 complex (Figure 4E), the D1 domain of CD155 binds VP1, and VP2 of one protomer and VP3 of adjacent protomer at the capsid quasi 3-fold axis.219 In the PV capsid, C-terminal, GH, EF, BC loops, C β strand of VP1, B β strand, GH loop of VP3 and EF loop of VP2 occupy the CD155 binding site.
Poliovirus infection is rapid and remarkably efficient, releasing as high as 10,000 mature virions per infected cell at 6 hrs post-infection.229 Even though a rapid replication warrants the applicability of PV in oncovirotherapy, a counter mechanism must be in place to minimize the neurotoxicity associated with wild-type PV. The neuro-attenuated variant of PV, PVSRIPO has completed a Phase I dose-finding clinical study in patients with grade IV malignant glioma with no neurotoxicity reported.230
Seneca Valley Virus
Seneca Valley Virus is the only member of Senecavirus genus of the Picornaviridae family. The overall structure of SVV has an icosahedral symmetry and is comprised of a non-enveloped protein capsid harboring a positive-sense ssRNA genome of approximately 7.3 kb (Figure 4F).231 Similar to CV and PV, the SVV genome encodes seven non-structural proteins and four structural proteins. To date, the SVV strains have been classified into 3 clades,232–234 with the prototype SVV-001 being the sole member of clade I.
SVV cell entry is dependent on its cellular receptor: anthrax toxin receptor 1 (ANTXR1), also known as tumor endothelial marker 8 (TEM8).235 ANTXR1 is a type I transmembrane protein overexpressed in many types of cancers, but weakly expressed in healthy tissues.236 The role of ANTXR1 is unknown beyond its function as a toxin and virus receptor; indeed, ANTXR1 knockout mice exhibit no major phenotypic abnormalities.237 However, ANTXR1 blockade has been shown to decrease tumor angiogenesis and to potentiate an anti-tumor effect towards certain cancers.238 Our group identified the surface exposed loops on SVV-001 capsid exterior, BC loop, loop II of VP1, the “puff” loop of VP2, and the “knob” loop of VP3 which form the binding site for the extracellular domain of ANTXR1 (Figure 4G).239 Furthermore, we showed that SVV binding site on ANTXR1 is non-conserved in its paralogous receptor, ANTXR2, which is expressed in normal cells, thereby providing a structural basis for tumor specificity of SVV.240 SVV empty capsid binds ANTXR1, suggesting it may have potential as a vaccine or as virus-like particles for the development of tumor-targeted delivery of drugs.240
As suggested from both functional and structural studies, the tumor tropism of SVV-001 is attributed to receptor-mediated internalization of the virus, a phenomenon common to other oncolytic picornaviruses. However, a successful SVV-001 infection may also require an additional innate immune defect.235 SVV-001 in its native form provides several advantages for oncovirotherapy: the native virus is genetically stable and non-toxic to healthy tissues, it is safe and it homes to tumors when administered systemically and pre-existing immunity for SVV is rare.241 Several preclinical, Phase I/II clinical studies have demonstrated the anti-tumor potential, intratumoral replication and safety of SVV in treating solid tumors with neuroendocrine features.242,243
Conclusion
Oncolytic viruses (OVs) either have an inherent ability to successfully replicate in cancer cells or they have been modified to exploit de-regulated signaling pathways in tumors. Nevertheless, the attachment of OVs to specific receptors found in cancers plays a pivotal role in OV tumor cell entry, subsequent viral replication and cell lysis. However, the expression of these receptors varies in different cancers and also among individual patients. Furthermore, the presence of natural receptors of OVs in normal cells may pose a potential challenge when the virus is pathogenic in nature. Therefore, understanding the structural details concerning how OVs interact with their receptors can inform the development of more efficient-targeted therapies to exploit cognate receptors and to reduce off-target cytotoxicity. Additionally, oncovirotherapy is constantly facing the challenge of overcoming anti-viral immunity in cancer patients. In this case, the knowledge of OV-receptor interactions is necessary to modify the viral capsid or envelope proteins in order to bypass the immune response without impairing the ability to bind their cellular receptors.
Disclosure
Dr John T Poirier reports personal fees from Perceiver Pharmaceuticals LLC, outside the submitted work. In addition, Dr Poirier has a patent WO2017096201A1 licensed to Perceiver Pharmaceuticals, LLC. The authors report no other conflicts of interest in this work.
References
1. Peters C, Grandi P, Nigim F. Updates on oncolytic virus immunotherapy for cancers. Mol Ther Oncolytics. 2019;12:259–262. doi:10.1016/j.omto.2019.01.008
2. Geletneky K, Hajda J, Angelova AL, et al. Oncolytic H-1 parvovirus shows safety and signs of immunogenic activity in a first phase I/Iia glioblastoma trial. Mol Ther. 2017;25(12):2620–2634. doi:10.1016/j.ymthe.2017.08.016
3. Aref S, Bailey K, Fielding A. Measles to the rescue: a review of oncolytic measles virus. Viruses. 2016;8(10):294.
4. Lazar I, Yaacov B, Shiloach T, et al. The oncolytic activity of newcastle disease virus NDV-HUJ on chemoresistant primary melanoma cells is dependent on the proapoptotic activity of the inhibitor of apoptosis protein livin. J Virol. 2010;84(1):639.
5. Hiley CT, Yuan M, Lemoine NR, Wang Y. Lister strain vaccinia virus, a potential therapeutic vector targeting hypoxic tumours. Gene Ther. 2009;17:281.
6. McCarthy C, Jayawardena N, Burga LN, Bostina M. Developing picornaviruses for cancer therapy. Cancers. 2019;11:5.
7. Conry RM, Westbrook B, McKee S, Norwood TG. Talimogene laherparepvec: first in class oncolytic virotherapy. Hum Vaccin Immunother. 2018;14(4):839–846.
8. Holl EK, Brown MC, Boczkowski D, et al. Recombinant oncolytic poliovirus, PVSRIPO, has potent cytotoxic and innate inflammatory effects, mediating therapy in human breast and prostate cancer xenograft models. Oncotarget. 2016;7(48):79828–79841.
9. Nemunaitis J, Ganly I, Khuri F, Arseneau J, Kuhn J, McCarty T. Selective replication and oncolysis in p53 mutant tumors with ONYX-015, an E1B-55kD gene-deleted adenovirus, in patients with advanced head and neck cancer: a phase II trial. Cancer Res. 2000;60:6359–66.
10. Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov. 2015;14:642. doi:10.1038/nrd4663
11. Breitbach CJ, De Silva NS, Falls TJ, et al. Targeting tumor vasculature with an oncolytic virus. Mol Ther. 2011;19(5):886–894. doi:10.1038/mt.2011.26
12. Vähä-Koskela M, Hinkkanen A. Tumor restrictions to oncolytic virus. Biomedicines. 2014;2(2):163–194. doi:10.3390/biomedicines2020163
13. Filley AC, Dey M. Immune system, friend or foe of oncolytic virotherapy? Front Oncol. 2017;7:106. doi:10.3389/fonc.2017.00106
14. Anderson DB, Laquerre S, Ghosh K, et al. Pseudotyping of glycoprotein D-deficient herpes simplex virus type 1 with vesicular stomatitis virus glycoprotein g enables mutant virus attachment and entry. J Virol. 2000;74(16):7698. doi:10.1128/JVI.74.16.7698-7698.2000
15. Ilett EJ, Bárcena M, Errington-Mais F, et al. Internalization of oncolytic reovirus by human dendritic cell carriers protects the virus from neutralization. Clin Cancer Res. 2011;17(9):2767. doi:10.1158/1078-0432.CCR-10-3266
16. Boonstra MC, de Geus SWL, Prevoo HAJM, et al. Selecting targets for tumor imaging: an overview of cancer-associated membrane proteins. Biomark Cancer. 2016;8:119–133. doi:10.4137/BIC.S38542
17. Pellet PE and Roizman B. The family herpesviridae: a brief introduction. In: Knipe DM and Howley PM, editors. In Fields Virology. Philadelphia [PA]: Lippincott Williams & Wilkins; 2007:2479–2499
18. Yuan S, Wang J, Zhu D, et al. Cryo-EM structure of a herpesvirus capsid at 3.1 Å. Science. 2018;360(6384):eaao7283. doi:10.1126/science.aao7283
19. Whitley RJ, Roizman B. Herpes simplex virus infections. Lancet. 2001;357(9267):1513–1518. doi:10.1016/S0140-6736(00)04638-9
20. Greig SL. Talimogene laherparepvec: first global approval. Drugs. 2016;76(1):147–154. doi:10.1007/s40265-015-0522-7
21. Andtbacka RHI, Collichio FA, Amatruda T, et al. Final planned overall survival (OS) from OPTiM, a randomized Phase III trial of talimogene laherparepvec (T-VEC) versus GM-CSF for the treatment of unresected stage IIIB/C/IV melanoma (NCT00769704). J Immunother Cancer. 2014;2(3):P263.
22. Andtbacka RHI, Curti B, Hallmeyer S, et al. Phase II CALM extension study: enhanced immune-cell infiltration within the tumour micro-environment of patients with advanced melanoma following intralesional delivery of Coxsackievirus A21. Eur J Cancer. 2015;51:S677–S677.
23. Andtbacka RHI, Kaufman HL, Collichio F, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33(25):2780–2788.
24. O’Donoghue C, Doepker MP, Zager JS. Talimogene laherparepvec: overview, combination therapy and current practices. Melanoma Manage. 2016;3(4):267–272.
25. Senior M. Checkpoint inhibitors go viral. Nat Biotechnol. 2019;37:12.
26. Markert JM, Razdan SN, Kuo H-C, et al. A phase 1 trial of oncolytic HSV-1, G207, given in combination with radiation for recurrent GBM demonstrates safety and radiographic responses. Mol Ther. 2014;22(5):1048–1055.
27. Geevarghese SK, Geller DA, de Haan HA, et al. Phase I/II study of oncolytic herpes simplex virus NV1020 in patients with extensively pretreated refractory colorectal cancer metastatic to the liver. Hum Gene Ther. 2010;21(9):1119–1128.
28. Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108(4):503–510.
29. Agelidis AM, Shukla D. Cell entry mechanisms of HSV: what we have learned in recent years. Future Virol. 2015;10(10):1145–1154.
30. Koelle DM, Corey L. Recent progress in herpes simplex virus immunobiology and vaccine research. Clin Microbiol Rev. 2003;16(1):96.
31. Di Giovine P, Settembre EC, Bhargava AK, et al. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog. 2011;7(9):e1002277.
32. Zhang N, Yan J, Lu G, et al. Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion. Nat Commun. 2011;2:577.
33. Carfı́ A, Willis SH, Whitbeck JC, et al. Herpes simplex virus glycoprotein d bound to the human receptor HveA. Mol Cell. 2001;8(1):169–179.
34. Kuroki K, Wang J, Ose T, et al. Structural basis for simultaneous recognition of an O-glycan and its attached peptide of mucin family by immune receptor PILRα. Proc Natl Acad Sci. 2014;111(24):8877.
35. Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol. 2009;83(13):6610–6623.
36. Malissen N, Macagno N, Granjeaud S, et al. HVEM: A novel cosignaling molecule of major interest in melanoma. J Clin Oncol. 2017;35(15_suppl):e14591–e14591.
37. Hokuto D, Sho M, Yamato I, et al. Clinical impact of herpesvirus entry mediator expression in human hepatocellular carcinoma. Eur J Cancer. 2015;51(2):157–165.
38. Lan X, Li S, Gao H, et al. Increased BTLA and HVEM in gastric cancer are associated with progression and poor prognosis. Onco Targets Ther. 2017;10:919–926.
39. Spear PG, Eisenberg RJ, Cohen GH. Three classes of cell surface receptors for alphaherpesvirus entry. Virology. 2000;275(1):1–8.
40. Ogita H, Takai Y. Nectins and nectin-like molecules: roles in cell adhesion, polarization, movement, and proliferation. IUBMB Life. 2006;58(5‐6):334–343.
41. Martin TA, Lane J, Harrison GM, Jiang WG. The expression of the Nectin complex in human breast cancer and the role of Nectin-3 in the control of tight junctions during metastasis. PLoS One. 2013;8(12):e82696–e82696.
42. Yu Z, Chan M-K, O-Charoenrat P, et al. Enhanced nectin-1 expression and herpes oncolytic sensitivity in highly migratory and invasive carcinoma. Clin Cancer Res. 2005;11(13):4889.
43. Yu Z, Adusumilli PS, Eisenberg DP, et al. Nectin-1 expression by squamous cell carcinoma is a predictor of herpes oncolytic sensitivity. Mol Ther. 2007;15(1):103–113.
44. Tampakis A, Tampaki EC, Nonni A, et al. Nectin-1 expression in colorectal cancer: is there a group of patients with high risk for early disease recurrence? Oncology. 2019;96(6):318–325.
45. Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol. 1998;72(12):9992–10002.
46. Nakano K, Asano R, Tsumoto K, et al. Herpes simplex virus targeting to the EGF receptor by a gD-specific soluble bridging molecule. Mol Ther. 2005;11(4):617–626.
47. Smith GL, Murphy BJ, Law M. Vaccinia virus motility. Annu Rev Microbiol. 2003;57(1):323–342.
48. Wilkinson L. Jenner’s smallpox vaccine. The riddle of vaccinia virus and its origin. Med Hist. 1982;26(1):94–95.
49. Joklik WK, Becker Y. The replication and coating of vaccinia DNA. J Mol Biol. 1964;10(3):452–474.
50. Chung C-S, Chen C-H, Ho M-Y, Huang C-Y, Liao C-L, Chang W. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J Virol. 2006;80(5):2127.
51. Ichihashi Y, Oie M. Neutralizing epitope on penetration protein of vaccinia virus. Virology. 1996;220(2):491–494.
52. Strassburg MA. The global eradication of smallpox. Am J Infect Control. 1982;10(2):53–59.
53. Guo ZS, Bartlett DL. Vaccinia as a vector for gene delivery. Expert Opin Biol Ther. 2004;4(6):901–917.
54. Haddad D. Genetically engineered vaccinia viruses as agents for cancer treatment, imaging, and transgene delivery. Front Oncol. 2017;7:96.
55. Thorne SH, Hwang TH, Kirn DH. Vaccinia virus and oncolytic virotherapy of cancer. Curr Opin Mol Ther. 2005;7(4):359–365.
56. Breitbach CJ, Burke J, Jonker D, et al. Intravenous delivery of a multi-mechanistic cancer-targeted oncolytic poxvirus in humans. Nature. 2011;477:99.
57. Heo J, Reid T, Ruo L, et al. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat Med. 2013;19:329.
58. Townsley AC, Weisberg AS, Wagenaar TR, Moss B. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J Virol. 2006;80(18):8899.
59. Carter GC, Law M, Hollinshead M, Smith GL. Entry of the vaccinia virus intracellular mature virion and its interactions with glycosaminoglycans. J Gen Virol. 2005;86(5):1279–1290.
60. Belting M. Glycosaminoglycans in cancer treatment. Thromb Res. 2014;133:S95–S101.
61. Rodriguez JF, Smith GL. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 1990;18(18):5347–5351.
62. Lin C-L, Chung C-S, Heine HG, Chang W. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J Virol. 2000;74(7):3353.
63. Hsiao J-C, Chung C-S, Chang W. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J Virol. 1998;72(10):8374.
64. Hsiao J-C, Chung C-S, Chang W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J Virol. 1999;73(10):8750.
65. Chung C-S, Hsiao J-C, Chang Y-S, Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J Virol. 1998;72(2):1577.
66. McCart JA, Ward JM, Lee J, et al. Systemic cancer therapy with a tumor-selective vaccinia virus mutant lacking thymidine kinase and vaccinia growth factor genes. Cancer Res. 2001;61(24):8751.
67. Gammon DB, Gowrishankar B, Duraffour S, Andrei G, Upton C, Evans DH. Vaccinia virus–encoded ribonucleotide reductase subunits are differentially required for replication and pathogenesis. PLoS Pathog. 2010;6(7):e1000984.
68. Banerjee AK. Transcription and replication of rhabdoviruses. Microbiol Rev. 1987;51(1):66–87.
69. Leyrat C, Ribeiro EA, FCA G, Ivanov I, Ruigrok RWH, Jamin M. Structure, interactions with host cell and functions of rhabdovirus phosphoprotein. Future Virol. 2011;6(4):465–481.
70. Felt SA, Grdzelishvili VZ. Recent advances in vesicular stomatitis virus-based oncolytic virotherapy: a 5-year update. J Gen Virol. 2017;98(12):2895–2911.
71. Hastie E, Grdzelishvili VZ. Vesicular stomatitis virus as a flexible platform for oncolytic virotherapy against cancer. J Gen Virol. 2012;93(12):2529–2545.
72. Stojdl DF, Lichty BD, tenOever BR, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4(4):263–275.
73. Finkelshtein D, Werman A, Novick D, Barak S, Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci. 2013;110(18):7306.
74. Gonias SL, Karimi-Mostowfi N, Murray SS, Mantuano E, Gilder AS. Expression of LDL receptor-related proteins (LRPs) in common solid malignancies correlates with patient survival. PLoS One. 2017;12(10):e0186649–e0186649.
75. Lillis AP, Van Duyn LB, Murphy-Ullrich JE, Strickland DK. LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev. 2008;88(3):887–918.
76. Nikolic J, Belot L, Raux H, et al. Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein. Nat Commun. 2018;9(1):1029.
77. Sieczkarski SB, Whittaker GR. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J Virol. 2002;76(20):10455.
78. Sun X, Roth SL, Bialecki MA, Whittaker GR. Internalization and fusion mechanism of vesicular stomatitis virus and related rhabdoviruses. Future Virol. 2010;5(1):85–96.
79. Le Boeuf F, Gebremeskel S, McMullen N, et al. Reovirus FAST protein enhances vesicular stomatitis virus oncolytic virotherapy in primary and metastatic tumor models. Mol Ther Oncolytics. 2017;6:80–89.
80. Salsman J, Top D, Boutilier J, Duncan R. Extensive syncytium formation mediated by the reovirus FAST proteins triggers apoptosis-induced membrane instability. J Virol. 2005;79(13):8090–8100.
81. Muik A, Stubbert LJ, Jahedi RZ, et al. Re-engineering vesicular stomatitis virus to abrogate neurotoxicity, circumvent humoral immunity, and enhance oncolytic potency. Cancer Res. 2014;74(13):3567.
82. Smelt SC, Borrow P, Kunz S, et al. Differences in affinity of binding of lymphocytic choriomeningitis virus strains to the cellular receptor α-dystroglycan correlate with viral tropism and disease kinetics. J Virol. 2001;75(1):448.
83. Kunz S, Rojek JM, Kanagawa M, et al. Posttranslational modification of α-dystroglycan, the cellular receptor for arenaviruses, by the glycosyltransferase LARGE is critical for virus binding. J Virol. 2005;79(22):14282.
84. Calogero A, Pavoni E, Gramaglia T, et al. Altered expression of alpha-dystroglycan subunit in human gliomas. Cancer Biol Ther. 2006;5(4):441–448.
85. Tong JG, Valdes YR, Sivapragasam M, et al. Spatial and temporal epithelial ovarian cancer cell heterogeneity impacts Maraba virus oncolytic potential. BMC Cancer. 2017;17(1):594.
86. Tong JG, Valdes YR, Barrett JW, et al. Evidence for differential viral oncolytic efficacy in an in vitro model of epithelial ovarian cancer metastasis. Mol Ther Oncolytics. 2015;2:15013.
87. Brun J, McManus D, Lefebvre C, et al. Identification of genetically modified maraba virus as an oncolytic rhabdovirus. Mol Ther. 2010;18(8):1440–1449.
88. Le Boeuf F, Selman M, Son HH, et al. Oncolytic maraba virus MG1 as a treatment for sarcoma. Int J Cancer. 2017;141(6):1257–1264.
89. Pol JG, Atherton MJ, Bridle BW, et al. Development and applications of oncolytic Maraba virus vaccines. Oncolytic Virotherapy. 2018;7: 117–128.
90. Brown VR, Bevins SN. A review of virulent Newcastle disease viruses in the United States and the role of wild birds in viral persistence and spread. Vet Res. 2017;48(1):68.
91. Bulbule NR. Virulence of Newcastle disease virus and diagnostic challenges. Adv Anim Vet Sci. 2015;3(5s):14–21.
92. Yurchenko KS, Zhou P, Kovner AV, Zavjalov EL, Shestopalova LV, Shestopalov AM. Oncolytic effect of wild-type Newcastle disease virus isolates in cancer cell lines in vitro and in vivo on xenograft model. PLoS One. 2018;13(4):e0195425.
93. Buijs PRA, van Eijck CHJ, Hofland LJ, Fouchier RAM. van den Hoogen BG. Different responses of human pancreatic adenocarcinoma cell lines to oncolytic Newcastle disease virus infection. Cancer Gene Ther. 2014;21:24.
94. Lam HY, Yeap SK, Rasoli M, et al. Safety and clinical usage of Newcastle disease virus in cancer therapy. Biomed Res Int. 2011;2011:718710.
95. Fournier P, Bian H, Szeberényi J, Schirrmacher V. Analysis of three properties of newcastle disease virus for fighting cancer: tumor-selective replication, antitumor cytotoxicity, and immunostimulation. In: Kirn DH, Liu T-C, Thorne SH, editors. Oncolytic Viruses: Methods and Protocols. Totowa, NJ: Humana Press; 2012:177–204.
96. Zaitsev V, von Itzstein M, Groves D, et al. Second sialic acid binding site in newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J Virol. 2004;78(7):3733.
97. Teoh ST, Ogrodzinski MP, Ross C, Hunter KW, Lunt SY. Sialic acid metabolism: a key player in breast cancer metastasis revealed by metabolomics. Front Oncol. 2018;8:174.
98. Huang Z, Panda A, Elankumaran S, Govindarajan D, Rockemann DD, Samal SK. The hemagglutinin-neuraminidase protein of newcastle disease virus determines tropism and virulence. J Virol. 2004;78(8):4176.
99. Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068.
100. Iorio RM, Field GM, Sauvron JM, et al. Structural and functional relationship between the receptor recognition and neuraminidase activities of the newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J Virol. 2001;75(4):1918. doi:10.1128/JVI.75.4.1918-1927.2001
101. Bousse TL, Taylor G, Krishnamurthy S, Portner A, Samal SK, Takimoto T. Biological significance of the second receptor binding site of newcastle disease virus hemagglutinin-neuraminidase protein. J Virol. 2004;78(23):13351. doi:10.1128/JVI.78.23.13351-13355.2004
102. Mahon PJ, Mirza AM, Iorio RM. Role of the two sialic acid binding sites on the newcastle disease virus HN protein in triggering the interaction with the F protein required for the promotion of fusion. J Virol. 2011;85(22):12079–12082. doi:10.1128/JVI.05679-11
103. Plattet P, Alves L, Herren M, Aguilar HC. Measles virus fusion protein: structure, function and inhibition. Viruses. 2016;8(4):112. doi:10.3390/v8040112
104. Plemper RK, Hammond AL. Synergizing vaccinations with therapeutics for measles eradication. Expert Opin Drug Discov. 2014;9(2):201–214. doi:10.1517/17460441.2014.867324
105. Gutsche I, Desfosses A, Effantin G, et al. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science. 2015;348(6235):704. doi:10.1126/science.aaa5139
106. Hashiguchi T, Kajikawa M, Maita N, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci. 2007;104(49):19535. doi:10.1073/pnas.0707830104
107. Hashiguchi T, Fukuda Y, Matsuoka R, et al. Structures of the prefusion form of measles virus fusion protein in complex with inhibitors. Proc Natl Acad Sci. 2018;115(10):2496. doi:10.1073/pnas.1718957115
108. Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol. 2009;17:124. doi:10.1038/nsmb.1726
109. Hashiguchi T, Ose T, Kubota M, et al. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol. 2011;18:135. doi:10.1038/nsmb.1969
110. Zhang X, Lu G, Qi J, et al. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat Struct Mol Biol. 2012;20:67. doi:10.1038/nsmb.2432
111. Anderson BD, Nakamura T, Russell SJ, Peng K-W. High CD46 receptor density determines preferential killing of tumor cells by oncolytic measles virus. Cancer Res. 2004;64(14):4919. doi:10.1158/0008-5472.CAN-04-0884
112. Nishiwada S, Sho M, Yasuda S, et al. Nectin-4 expression contributes to tumor proliferation, angiogenesis and patient prognosis in human pancreatic cancer. J Exp Clin Cancer Res. 2015;34(1):30. doi:10.1186/s13046-015-0144-7
113. Tatsuo H, Ono N, Tanaka K, SLAM YY. (CDw150) is a cellular receptor for measles virus. Nature. 2000;406(6798):893–897. doi:10.1038/35022579
114. Russell S. CD46: A complement regulator and pathogen receptor that mediates links between innate and acquired immune function. Tissue Antigens. 2004;64(2):111–118. doi:10.1111/j.1399-0039.2004.00277.x
115. Persson BD, Reiter DM, Marttila M, et al. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat Struct Mol Biol. 2007;14:164. doi:10.1038/nsmb1190
116. Santiago C, Björling E, Stehle T, Casasnovas JM. Distinct kinetics for binding of the CD46 and SLAM receptors to overlapping sites in the measles virus hemagglutinin protein. J Biol Chem. 2002;277(35):32294–32301. doi:10.1074/jbc.M202973200
117. Lecouturier V, Fayolle J, Caballero M, et al. Identification of two amino acids in the hemagglutinin glycoprotein of measles virus (MV) that govern hemadsorption, HeLa cell fusion, and CD46 downregulation: phenotypic markers that differentiate vaccine and wild-type MV strains. J Virol. 1996;70(7):4200.
118. Zhang S-C, Wang W-L, Cai W-S, Jiang K-L, Yuan Z-W. Engineered measles virus Edmonston strain used as a novel oncolytic viral system against human hepatoblastoma. BMC Cancer. 2012;12(1):427. doi:10.1186/1471-2407-12-427
119. Galanis E, Atherton PJ, Maurer MJ, et al. Oncolytic measles virus expressing the sodium iodide symporter to treat drug-resistant ovarian cancer. Cancer Res. 2015;75(1):22. doi:10.1158/0008-5472.CAN-14-3569
120. Galanis E, Hartmann LC, Cliby WA, et al. Phase I trial of intraperitoneal administration of an oncolytic measles virus strain engineered to express carcinoembryonic antigen for recurrent ovarian cancer. Cancer Res. 2010;70(3):875. doi:10.1158/0008-5472.CAN-09-2762
121. Russell SJ, Federspiel MJ, Peng K-W, et al. Remission of disseminated cancer after systemic oncolytic virotherapy. Mayo Clin Proc. 2014;89(7):926–933. doi:10.1016/j.mayocp.2014.04.003
122. Vongpunsawad S, Oezgun N, Braun W, Cattaneo R. Selectively receptor-blind measles viruses: identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J Virol. 2004;78(1):302–313. doi:10.1128/JVI.78.1.302-313.2004
123. Nakamura T, Peng K-W, Vongpunsawad S, et al. Antibody-targeted cell fusion. Nat Biotechnol. 2004;22(3):331–336. doi:10.1038/nbt942
124. Nakamura T, Peng K-W, Harvey M, et al. Rescue and propagation of fully retargeted oncolytic measles viruses. Nat Biotechnol. 2005;23(2):209–214. doi:10.1038/nbt1060
125. Buckwalter SP, Teo R, Espy MJ, Sloan LM, Smith TF, Pritt BS. Real-time qualitative PCR for 57 human adenovirus types from multiple specimen sources. J Clin Microbiol. 2012;50(3):766. doi:10.1128/JCM.01263-11
126. Cheng L, Huang X, Li X, et al. Cryo-EM structures of two bovine adenovirus type 3 intermediates. Virology. 2014;450–451:174–181. doi:10.1016/j.virol.2013.12.012
127. Ghebremedhin B. Human adenovirus: viral pathogen with increasing importance. Eur J Microbiol Immunol (Bp). 2014;4(1):26–33. doi:10.1556/EuJMI.4.2014.1.2
128. Yabe Y, Samper L, Bryan E, Taylor G, Trentin JJ. Oncogenic effect of human adenovirus type 12, in mice. Science. 1964;143(3601):46. doi:10.1126/science.143.3601.46
129. Trentin JJ, Yabe Y, Taylor G. The quest for human cancer viruses. Science. 1962;137(3533):835. doi:10.1126/science.137.3530.571
130. Niemann J, Kühnel F. Oncolytic viruses: adenoviruses. Virus Genes. 2017;53(5):700–706. doi:10.1007/s11262-017-1488-1
131. Khuri FR, Nemunaitis J, Ganly I, et al. A controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat Med. 2000;6:879. doi:10.1038/78638
132. Lang FF, Conrad C, Gomez-Manzano C, et al. Phase I study of DNX-2401 (Delta-24-RGD) oncolytic adenovirus: replication and immunotherapeutic effects in recurrent malignant glioma. J Clin Oncol. 2018;36(14):1419–1427. doi:10.1200/JCO.2017.75.8219
133. Sharma A, Li X, Bangari DS, Mittal SK. Adenovirus receptors and their implications in gene delivery. Virus Res. 2009;143(2):184–194. doi:10.1016/j.virusres.2009.02.010
134. Leopold PL, Crystal RG. Intracellular trafficking of adenovirus: many means to many ends. Adv Drug Deliv Rev. 2007;59(8):810–821. doi:10.1016/j.addr.2007.06.007
135. Zhang Y, Bergelson JM. Adenovirus Receptors. J Virol. 2005;79(19):12125. doi:10.1128/JVI.79.20.12905-12913.2005
136. Chrétien I, Marcuz A, Courtet M, et al. CTX, a Xenopus thymocyte receptor, defines a molecular family conserved throughout vertebrates. Eur J Immunol. 1998;28(12):4094–4104. doi:10.1002/(SICI)1521-4141(199812)28:12<4094::AID-IMMU4094>3.0.CO;2-2
137. Freimuth P, Springer K, Berard C, Hainfeld J, Bewley M, Flanagan J. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J Virol. 1999;73(2):1392.
138. Kirby I, Davison E, Beavil AJ, et al. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J Virol. 2000;74(6):2804. doi:10.1128/JVI.74.6.2804-2813.2000
139. Reeh M, Bockhorn M, Görgens D, et al. Presence of the Coxsackievirus and Adenovirus Receptor (CAR) in human neoplasms: a multitumour array analysis. Br J Cancer. 2013;109:1848. doi:10.1038/bjc.2013.623
140. Küster K, Koschel A, Rohwer N, Fischer A, Wiedenmann B, Anders M. Downregulation of the coxsackie and adenovirus receptor in cancer cells by hypoxia depends on HIF-1α. Cancer Gene Ther. 2009;17:141. doi:10.1038/cgt.2009.49
141. Wu PC, Wang Q, Dong ZM, et al. Expression of coxsackie and adenovirus receptor distinguishes transitional cancer states in therapy-induced cellular senescence. Cell Death Dis. 2010;1:e70. doi:10.1038/cddis.2010.47
142. Fishelson Z, Donin N, Zell S, Schultz S, Kirschfink M. Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Mol Immunol. 2003;40(2):109–123. doi:10.1016/s0161-5890(03)00112-3
143. Do M-H, To PK, Cho Y-S, et al. Targeting CD46 enhances anti-tumoral activity of adenovirus type 5 for bladder cancer. Int J Mol Sci. 2018;19(9):2694. doi:10.3390/ijms19092694
144. Lenman A, Liaci AM, Liu Y, et al. Polysialic acid is a cellular receptor for human adenovirus 52. Proc Natl Acad Sci. 2018;115(18):E4264. doi:10.1073/pnas.1716900115
145. Petridis AK, Wedderkopp H, Hugo HH, Maximilian Mehdorn H. Polysialic acid overexpression in malignant astrocytomas. Acta Neurochir (Wien). 2009;151(6):601–604. doi:10.1007/s00701-009-0324-3
146. Amoureux M-C, Coulibaly B, Chinot O, et al. Polysialic Acid Neural Cell Adhesion Molecule (PSA-NCAM) is an adverse prognosis factor in glioblastoma, and regulates olig2 expression in glioma cell lines. BMC Cancer. 2010;10(1):91. doi:10.1186/1471-2407-10-663
147. Suzuki M, Suzuki M, Nakayama J, et al. Polysialic acid facilitates tumor invasion by glioma cells. Glycobiology. 2005;15(9):887–894. doi:10.1093/glycob/cwi071
148. Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2004;23(1):515–548. doi:10.1146/annurev.immunol.23.021704.115611
149. Short JJ, Pereboev AV, Kawakami Y, Vasu C, Holterman MJ, Curiel DT. Adenovirus serotype 3 utilizes CD80 (B7.1) and CD86 (B7.2) as cellular attachment receptors. Virology. 2004;322(2):349–359. doi:10.1016/j.virol.2004.02.016
150. Tatsis N, Ertl HCJ. Adenoviruses as vaccine vectors. Mol Ther. 2004;10(4):616–629. doi:10.1016/j.ymthe.2004.07.013
151. Wang H, Li Z-Y, Liu Y, et al. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat Med. 2010;17:96. doi:10.1038/nm.2270
152. Chitaev NA, Troyanovsky SM. Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein and desmocollin, contributes to cell-cell adhesion. J Cell Biol. 1997;138(1):193. doi:10.1083/jcb.138.1.193
153. S-E-H S, Trinnaman B, Martin S, Major S, Hutchinson J, Magee AI. Molecular interactions between desmosomal cadherins. Biochem J. 2002;362(2):317. doi:10.1042/bj3620317
154. Harrison OJ, Brasch J, Lasso G, et al. Structural basis of adhesive binding by desmocollins and desmogleins. Proc Natl Acad Sci. 2016;113(26):7160. doi:10.1073/pnas.1606272113
155. Biedermann K, Vogelsang H, Becker I, et al. Desmoglein 2 is expressed abnormally rather than mutated in familial and sporadic gastric cancer. J Pathol. 2005;207(2):199–206. doi:10.1002/path.1821
156. Schmitt CJ, Franke WW, Goerdt S, Falkowska-Hansen B, Rickelt S, Peitsch WK. Homo- and heterotypic cell contacts in malignant melanoma cells and desmoglein 2 as a novel solitary surface glycoprotein. J Investig Dermatol. 2007;127(9):2191–2206. doi:10.1038/sj.jid.5700849
157. Abbod MF, Hamdy FC, Linkens DA, Catto JWF. Predictive modeling in cancer: where systems biology meets the stock market. Expert Rev Anticancer Ther. 2009;9(7):867–870. doi:10.1586/era.09.47
158. Cai F, Zhu Q, Miao Y, Shen S, Su X, Shi Y. Desmoglein-2 is overexpressed in non-small cell lung cancer tissues and its knockdown suppresses NSCLC growth by regulation of p27 and CDK2. J Cancer Res Clin Oncol. 2017;143(1):59–69. doi:10.1007/s00432-016-2250-0
159. Vassal-Stermann E, Effantin G, Zubieta C, et al. CryoEM structure of adenovirus type 3 fibre with desmoglein 2 shows an unusual mode of receptor engagement. Nat Commun. 2019;10(1):1181. doi:10.1038/s41467-019-09220-y
160. Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell. 1993;73(2):309–319. doi:10.1016/0092-8674(93)90231-E
161. Chiu CY, Mathias P, Nemerow GR, Stewart PL. Structure of adenovirus complexed with its internalization receptor, alphavbeta5 integrin. J Virol. 1999;73(8):6759–6768.
162. Veesler D, Cupelli K, Burger M, Gräber P, Stehle T, Johnson JE. Single-particle EM reveals plasticity of interactions between the adenovirus penton base and integrin αVβ<sub>3</sub>. Proc Natl Acad Sci. 2014;111(24):8815.
163. Bai M, Harfe B, Freimuth P. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J Virol. 1993;67(9):5198.
164. Freimuth P. A human cell line selected for resistance to adenovirus infection has reduced levels of the virus receptor. J Virol. 1996;70(6):4081.
165. Dmitriev I, Krasnykh V, Miller CR, et al. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J Virol. 1998;72(12):9706–9713.
166. Millward S, Graham AF. Structural studies on reovirus: discontinuities in the genome. Proc Natl Acad Sci U S A. 1970;65(2):422–429. doi:10.1073/pnas.65.2.422
167. Loh PC, Shatkin AJ. Structural proteins of reoviruses. J Virol. 1968;2(11):1353.
168. Chappell JD, Prota AE, Dermody TS, Stehle T. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. Embo J. 2002;21(1–2):1. doi:10.1093/emboj/21.1.1
169. Kirchner E, Guglielmi KM, Strauss HM, Dermody TS, Stehle T. Structure of reovirus σ1 in complex with its receptor junctional adhesion molecule-a. PLoS Pathog. 2008;4(12):e1000235. doi:10.1371/journal.ppat.1000235
170. Reiter DM, Frierson JM, Halvorson EE, Kobayashi T, Dermody TS, Stehle T. Crystal structure of reovirus attachment protein σ1 in complex with sialylated oligosaccharides. PLoS Pathog. 2011;7(8):e1002166. doi:10.1371/journal.ppat.1002166
171. Naik MU, Naik TU, Suckow AT, Duncan MK, Naik UP. Attenuation of junctional adhesion molecule-A is a contributing factor for breast cancer cell invasion. Cancer Res. 2008;68(7):2194. doi:10.1158/0008-5472.CAN-07-3057
172. Phillips MB, Stuart JD, Stewart RMR, Berry JTL, Mainou BA, Boehme KW. Current understanding of reovirus oncolysis mechanisms. Oncolytic Virotherapy. 2018. doi:10.2147/OV.S143808
173. Gollamudi R, Ghalib MH, Desai KK, et al. Intravenous administration of Reolysin®, a live replication competent RNA virus is safe in patients with advanced solid tumors. Invest New Drugs. 2010;28(5):641–649. doi:10.1007/s10637-009-9279-8
174. Morris DG, Feng X, DiFrancesco LM, et al. REO-001: A phase I trial of percutaneous intralesional administration of reovirus type 3 dearing (Reolysin®) in patients with advanced solid tumors. Invest New Drugs. 2013;31(3):696–706. doi:10.1007/s10637-012-9865-z
175. Vidal L, Pandha HS, Yap TA, et al. A phase I study of intravenous oncolytic reovirus type 3 Dearing in patients with advanced cancer. Clin Cancer Res. 2008;14:7127–7137.
176. Noonan AM, Farren MR, Geyer SM, et al. Randomized phase 2 trial of the oncolytic virus pelareorep (Reolysin) in upfront treatment of metastatic pancreatic adenocarcinoma. Mol Ther. 2016;24(6):1150–1158. doi:10.1038/mt.2016.66
177. Comins C, Spicer J, Protheroe A, et al. REO-10: a phase i study of intravenous reovirus and docetaxel in patients with advanced cancer. Clin Cancer Res. 2010;16(22):5564. doi:10.1158/1078-0432.CCR-10-0613
178. Anderson MJ, Jones SE, Fisher-Hoch SP, et al. Human parvovirus, the cause of erythema infectiosum (FIFTH DISEASE)? Lancet. 1983;321(8338):1378. doi:10.1016/S0140-6736(83)92152-9
179. Agbandje M, Kajigaya S, McKenna R, Young NS, Rossmann MG. The structure of human parvovirus B19 at 8 Å; resolution. Virology. 1994;203(1):106–115. doi:10.1006/viro.1994.1460
180. Marchini A, Bonifati S, Scott EM, Angelova AL, Rommelaere J. Oncolytic parvoviruses: from basic virology to clinical applications. Virol J. 2015;12:6. doi:10.1186/s12985-014-0223-y
181. Brown KE, Anderson SM, Young NS. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262(5130):114. doi:10.1126/science.8211117
182. Weigel-Kelley KA, Yoder MC, Srivastava A. α5β1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of β1 integrin for viral entry. Blood. 2003;102(12):3927. doi:10.1182/blood-2003-05-1522
183. Roman J, Ritzenthaler JD, Roser-Page S, Sun X, Han S. alpha5beta1-integrin expression is essential for tumor progression in experimental lung cancer. Am J Respir Cell Mol Biol. 2010;43(6):684–691. doi:10.1165/rcmb.2009-0375OC
184. Allaume X, El-Andaloussi N, Leuchs B, et al. Retargeting of rat parvovirus H-1PV to cancer cells through genetic engineering of the viral capsid. J Virol. 2012;86(7):3452. doi:10.1128/JVI.06208-11
185. Hafenstein S, Palermo LM, Kostyuchenko VA, et al. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc Natl Acad Sci. 2007;104(16):6585. doi:10.1073/pnas.0701574104
186. Shen Y, Li X, Dong D, Zhang B, Xue Y, Shang P. Transferrin receptor 1 in cancer: a new sight for cancer therapy. Am J Cancer Res. 2018;8(6):916–931.
187. Sin J, Mangale V, Thienphrapa W, Gottlieb RA, Feuer R. Recent progress in understanding coxsackievirus replication, dissemination, and pathogenesis. Virology. 2015;484:288–304. doi:10.1016/j.virol.2015.06.006
188. Oberste MS, Maher K, Pallansch MA. Molecular phylogeny and proposed classification of the simian picornaviruses. J Virol. 2002;76(3):1244. doi:10.1128/JVI.76.3.1244-1251.2002
189. Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229(4720):1358. doi:10.1126/science.229.4718.1038-a
190. Xu S-J, Yang H, Yao X-J, et al. Complete genome sequences of four coxsackievirus A16 strains isolated from four children with severe hand, foot, and mouth disease. Genome Announc. 2017;5(31):e00760–e00717. doi:10.1128/genomeA.00760-17
191. Zhu L, Sun Y, Fan J, et al. Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating. Nat Commun. 2018;9(1):4985. doi:10.1038/s41467-018-07531-0
192. Hogle JM. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu Rev Microbiol. 2002;56(1):677–702. doi:10.1146/annurev.micro.56.012302.160757
193. Bostina M, Levy H, Filman DJ, Hogle JM. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J Virol. 2011;85(2):776. doi:10.1128/JVI.00531-10
194. Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J Virol. 2013;87(7):3903. doi:10.1128/JVI.03209-12
195. Lyu K, Ding J, Han J-F, et al. Human enterovirus 71 uncoating captured at atomic resolution. J Virol. 2014;88(6):3114. doi:10.1128/JVI.03029-13
196. He Y, Chipman PR, Howitt J, et al. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat Struct Biol. 2001;8:874. doi:10.1038/nsb1001-874
197. Martino TA, Petric M, Weingartl H, et al. The Coxsackie-Adenovirus Receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group B and by swine vesicular disease virus. Virology. 2000;271(1):99–108. doi:10.1006/viro.2000.0324
198. Knowlton KU, Jeon ES, Berkley N, Wessely R, Huber S. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol. 1996;70(11):7811.
199. Yoder JD, Cifuente JO, Pan J, Bergelson JM, Hafenstein S. The crystal structure of a coxsackievirus B3-RD variant and a refined 9-angstrom cryo-electron microscopy reconstruction of the virus complexed with Decay-Accelerating Factor (DAF) provide a new footprint of DAF on the virus surface. J Virol. 2012;86(23):12571. doi:10.1128/JVI.01592-12
200. Goodfellow IG, Evans DJ, Blom AM, et al. Inhibition of coxsackie B virus infection by soluble forms of its receptors: binding affinities, altered particle formation, and competition with cellular receptors. J Virol. 2005;79(18):12016. doi:10.1128/JVI.79.20.12905-12913.2005
201. Xiao C, Bator-Kelly CM, Rieder E, et al. The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure. 2005;13(7):1019–1033. doi:10.1016/j.str.2005.04.011
202. Baggen J, Hurdiss DL, Zocher G, et al. Role of enhanced receptor engagement in the evolution of a pandemic acute hemorrhagic conjunctivitis virus. Proc Natl Acad Sci. 2018;115(2):397. doi:10.1073/pnas.1713284115
203. Shafren DR. Viral cell entry induced by cross-linked decay-accelerating factor. J Virol. 1998;72(11):9407.
204. Tsai S-T, Wang P-J, Liou N-J, Lin P-S, Chen C-H, Chang W-C. ICAM1 is a potential cancer stem cell marker of esophageal squamous cell carcinoma. PLoS One. 2015;10(11):e0142834. doi:10.1371/journal.pone.0142834
205. Kotteas EA, Boulas P, Gkiozos I, Tsagkouli S, Tsoukalas G, Syrigos KN. The intercellular cell adhesion molecule-1 (ICAM-1) in lung cancer: implications for disease progression and prognosis. Anticancer Res. 2014;34(9):4665–4672.
206. Benedicto A, Romayor I, Arteta B. Role of liver ICAM-1 in metastasis. Oncol Lett. 2017;14(4):3883–3892.
207. Inoue T, Yamakawa M, Takahashi T. Expression of complement regulating factors in gastric cancer cells. Mol Pathol. 2002;55(3):193.
208. Nakagawa M, Mizuno M, Kawada M, et al. Polymorphic expression of decay-accelerating factor in human colorectal cancer. J Gastroenterol Hepatol. 2001;16(2):184–189.
209. Bella J, Kolatkar PR, Marlor CW, Greve JM, Rossmann MG. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc Natl Acad Sci U S A. 1998;95(8):4140–4145.
210. Au GG, Lincz LF, Enno A, Shafren DR. Oncolytic coxsackievirus A21 as a novel therapy for multiple myeloma. Br J Haematol. 2007;137(2):133–141.
211. Skelding KA, Barry RD, Shafren DR. Enhanced oncolysis mediated by coxsackievirus A21 in combination with doxorubicin hydrochloride. Invest New Drugs. 2012;30(2):568–581.
212. Miyamoto S, Inoue H, Nakamura T, et al. Coxsackievirus B3 is an oncolytic virus with immunostimulatory properties that is active against lung adenocarcinoma. Cancer Res. 2012;72(10):2609–2621.
213. Melnick JL. Current status of poliovirus infections. Clin Microbiol Rev. 1996;9(3):293.
214. Yakovenko ML, Cherkasova EA, Rezapkin GV, et al. Antigenic evolution of vaccine-derived polioviruses: changes in individual epitopes and relative stability of the overall immunological properties. J Virol. 2006;80(6):2641.
215. Jiang P, Liu Y, Ma H-C, Paul AV, Wimmer E. Picornavirus Morphogenesis. Microbiol Mol Biol Rev. 2014;78(3):418.
216. Miller ST, Hogle JM, Filman DJ. Ab initio phasing of high-symmetry macromolecular complexes: successful phasing of authentic poliovirus data to 3.0 Å resolution11Edited by I. A. Wilson. J Mol Biol. 2001;307(2):499–512.
217. He Y, Mueller S, Chipman PR, et al. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J Virol. 2003;77(8):4827.
218. Zhang P, Mueller S, Morais MC, et al. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc Natl Acad Sci. 2008;105(47):18284.
219. Strauss M, Filman DJ, Belnap DM, Cheng N, Noel RT, Hogle JM. Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J Virol. 2015;89(8):4143.
220. He Y, Bowman VD, Mueller S, et al. Interaction of the poliovirus receptor with poliovirus. Proc Natl Acad Sci. 2000;97(1):79.
221. Butan C, Filman DJ, Hogle JM. Cryo-electron microscopy reconstruction shows poliovirus 135S particles poised for membrane interaction and RNA release. J Virol. 2014;88(3):1758.
222. Levy HC, Bostina M, Filman DJ, Hogle JM. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J Virol. 2010;84(9):4426.
223. Gao J, Zheng Q, Xin N, Wang W, Zhao C. CD155, an onco-immunologic molecule in human tumors. Cancer Sci. 2017;108(10):1934–1938.
224. Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56(5):855–865.
225. Chadéneau C, LeCabellec M, LeMoullac B, Meflah K, Denis MG. Over-expression of a novel member of the immunoglobulin superfamily in Min mouse intestinal adenomas. Int J Cancer. 1996;68(6):817–821.
226. Chadéneau C, LeMoullac B, Denis MG. A novel member of the immunoglobulin gene superfamily expressed in rat carcinoma cell lines. J Biol Chem. 1994;269(22):15601–15605.
227. Lim Y-P, Fowler LC, Hixson DC, Wehbe T, Thompson NL. TuAg.1 is the liver isoform of the rat colon tumor-associated antigen pE4 and a member of the immunoglobulin-like supergene family. Cancer Res. 1996;56(17):3934.
228. Erickson BM, Thompson NL, Hixson DC. Tightly regulated induction of the adhesion molecule necl-5/CD155 during rat liver regeneration and acute liver injury. Hepatology. 2006;43(2):325–334.
229. Bodian D. Histopathologic basis of clinical findings in poliomyelitis. Am J Med. 1949;6(5):563–578.
230. Desjardins A, Gromeier M, Herndon JE
231. Hales LM, Knowles NJ, Reddy PS, Xu L, Hay C, Hallenbeck PL. Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus. J Gen Virol. 2008;89(5):1265–1275.
232. Xu W, Hole K, Goolia M, et al. Genome wide analysis of the evolution of senecavirus A from swine clinical material and assembly yard environmental samples. PLoS One. 2017;12(5):e0176964.
233. Zhang X, Zhu Z, Yang F, et al. Review of seneca valley virus: a call for increased surveillance and research. Front Microbiol. 2018;9:940.
234. Leme RA, Alfieri AF, Alfieri AA. Update on senecavirus infection in pigs. Viruses. 2017;9(7):170.
235. Miles LA, Burga LN, Gardner EE, Bostina M, Poirier JT, Rudin CM. Anthrax toxin receptor 1 is the cellular receptor for Seneca Valley virus. J Clin Invest. 2017;127(8):2957–2967.
236. Bachran C, Leppla HS. Tumor targeting and drug delivery by anthrax toxin. Toxins. 2016;8(7):
237. Cullen M, Seaman S, Chaudhary A, et al. Host-derived tumor endothelial marker 8 promotes the growth of melanoma. Cancer Res. 2009;69(15):6021.
238. Chaudhary A, Hilton Mary B, Seaman S, et al. TEM8/ANTXR1 blockade inhibits pathological angiogenesis and potentiates tumoricidal responses against multiple cancer types. Cancer Cell. 2012;21(2):212–226.
239. Jayawardena N, Burga LN, Easingwood RA, Takizawa Y, Wolf M, Bostina M. Structural basis for anthrax toxin receptor 1 recognition by Seneca Valley Virus. Proc Natl Acad Sci. 2018;115(46):E10934.
240. Strauss M, Jayawardena N, Sun E, Easingwood RA, Burga LN, Bostina M. Cryo-electron microscopy structure of Seneca Valley Virus procapsid. J Virol. 2018;92(6):e01927–e01917.
241. Burke MJ. Oncolytic Seneca Valley Virus: past perspectives and future directions. Oncolytic Virotherapy. 2016;5:81–89.
242. Reddy PS, Burroughs KD, Hales LM, et al. Seneca Valley Virus, a systemically deliverable oncolytic picornavirus, and the treatment of neuroendocrine cancers. J Natl Cancer Inst. 2007;99(21):1623–1633.
243. Rudin CM, Poirier JT, Senzer NN, et al. Phase I clinical study of Seneca Valley Virus (SVV-001), a replication-competent picornavirus, in advanced solid tumors with neuroendocrine features. Clin Cancer Res. 2011;17(4):888–895.
244. Bender FC, Whitbeck JC, Lou H, Cohen GH, Eisenberg RJ. Herpes Simplex Virus Glycoprotein B Binds to Cell Surfaces Independently of Heparan Sulfate and Blocks Virus Entry. J Virol. 2005;79(18):11588–11597.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.