Back to Journals » Journal of Asthma and Allergy » Volume 14
Transcriptional Changes in Chronic Rhinosinusitis with Asthma Favor a Type 2 Molecular Endotype Independent of Polyp Status
Authors Gill AS, Pulsipher A , Sumsion JS, Oakley GM, Leclair LW, Howe H, Orlandi RR , Alt JA
Received 23 February 2021
Accepted for publication 16 March 2021
Published 21 April 2021 Volume 2021:14 Pages 405—413
DOI https://doi.org/10.2147/JAA.S301825
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Luis Garcia-Marcos
Amarbir S Gill, 1 Abigail Pulsipher, 1 Jorgen S Sumsion, 2 Gretchen M Oakley, 1 Laurie W Leclair, 3 Heather Howe, 3 Richard R Orlandi, 1 Jeremiah A Alt 1
1Division of Otolaryngology – Head and Neck Surgery, Department of Surgery, University of Utah, Salt Lake City, UT, USA; 2School of Medicine, University of Utah, Salt Lake, USA; 3Department of Pulmonology/Critical Care Medicine, University of Utah, Salt Lake City, UT, USA
Correspondence: Jeremiah A Alt
Division of Otolaryngology – Head & Neck Surgery, Department of Surgery, University of Utah School of Medicine, 50 North Medical Drive, Room 3C120, Salt Lake City, UT, 84132, USA
Tel +1 801 585-7143
Fax +1 801 585-5744
Email [email protected]
Background: Data regarding the inflammatory profile of patients with asthma and chronic rhinosinusitis (CRS-A) with (CRSwNP-A) and without (CRSsNP-A) nasal polyposis remain limited.
Objective: Define and compare systemic transcriptional changes in patients with CRS-A to those with non-asthma-related CRS with (CRSwNP) and without nasal polyposis (CRSsNP).
Methods: Thirty-four patients with CRS-A (n=19) and CRS (n=15) were prospectively enrolled into an observational study. Demographic information and subjective and objective disease severity measures were recorded. Multiplex gene expression analysis of mRNA extracted from peripheral blood was performed. A total of 594 genes associated with innate/adaptive immunity were analyzed using NanoString technology. Gene expression ratios were reported for genes that were differentially expressed among these cohorts. Linear regression analysis was used to compare the mRNA transcript copy numbers for each gene with disease severity.
Results: There was no significant difference in age, gender, nasal polyposis, or health-related quality of life measures between the two groups (p> 0.05). HLA class II histocompatibility antigen, DRB3-1 beta chain (HLA-DRB3) was significantly upregulated in the peripheral blood of patients with CRSsNP-A compared to CRSsNP, whereas chemokine (C-C motif) ligands 4 (CCL4) and zinc finger protein helios (IKZF2) were significantly upregulated in CRSwNP-A compared to CRSwNP (p< 0.05).
Conclusion: Patients with CRSsNP-A demonstrate a molecular endotype associated with a Th2-dominant inflammatory profile compared to CRSsNP. Patients with CRSwNP-A similarly demonstrate an overrepresentation of genes associated with Th2-driven inflammation compared to patients with CRSwNP.
Keywords: chronic rhinosinusitis, asthma, gene transcription, inflammatory markers, nasal polyposis, endotype
Corrigendum for this paper has been published
Introduction
Asthma, a disease of the large and small bronchi, affects over 26 million Americans,1 while chronic rhinosinusitis (CRS), a disease characterized by chronic inflammation of the sinonasal mucosa, affects over 28 million.2 Prevalence of both diseases is rising with no cure. A significant portion of patients with asthma develop comorbid CRS, and this particular phenotype of asthma can be among the most difficult to manage, demonstrating poorer outcomes and increased exacerbations and healthcare costs.3
CRS is subtyped by phenotype [ie, CRS without nasal polyposis (CRSsNP) vs CRS with nasal polyposis (CRSwNP)], as well as by molecular and inflammatory endotypes.4 These subtypes are important to consider as they provide us with some information regarding prognosis. CRSwNP is known to present with greater disease severity and higher recurrence rates compared to CRSsNP.4–6 Historically, CRSwNP has been more closely linked to asthma, with initial findings demonstrating a similar inflammatory milieu in patients with asthma and those with CRSwNP,4 having a cytokine profile heavily dominated by a type 2 (Th2 cell-mediated) inflammatory response.
In contrast, cytokine profiles typically associated with CRSsNP can present as a mixture of type 1 (Th1 cell-mediated), Th2, and type 3 (Th17 cell-mediated) inflammation.4,7 These inflammatory endotypes themselves are influenced by molecular endotypes and associated upstream transcriptional changes that occur in these disease cohorts.4 Despite this, there is currently very limited data on molecular endotypes among patients with CRS with asthma (CRS-A), with no studies examining this topic in patients with CRSsNP and asthma (CRSsNP-A).
Given the particular difficulty in treating patients with CRS-A and the lack of data outlining gene transcriptional changes in this cohort, the objective of this study was to define and compare the systemic molecular endotype of peripheral blood in patients with CRS-A to those with non-asthma-related CRS (CRS).
Methods
Thirty-four patients with CRS-A (n=19) and CRS (n=15) were enrolled into an observational study. Institutional Review Board (IRB) approval was obtained at the University of Utah. Written consent was obtained from patients undergoing treatment of CRSsNP and CRSwNP, as defined by the American Academy of Otolaryngology–Head and Neck Surgery (AAO-HNS),8 who were prospectively enrolled between 2013 and 2020. All patients were over the age of 18 and informed about the purpose of the study. This study was conducted in accordance with the Declaration of Helsinki. Inclusion criteria included a diagnosis of CRS and/or asthma; the latter was confirmed by physician diagnosis, asthma medication usage, or pulmonary function testing. Patients on oral steroids and/or the following diagnoses were excluded in order to decrease the potential non-CRS-associated contributions to systemic inflammation: allergic fungal rhinosinusitis, eosinophilic granulomatosis with polyangiitis, systemic lupus erythematosus, chronic obstructive pulmonary disease, multiple sclerosis, cystic fibrosis, cancer, and smoking and alcohol use. All recruited patients were included in the analysis.
Demographics
The following demographic information, clinical characteristics, and comorbidities were collected for each patient: age, gender, smoking status, polyp status, migraine, diabetes, gastroesophageal reflux disease, aspirin sensitivity, cystic fibrosis, and atopy.
Measures of Disease Severity
Endoscopic examination (Lund-Mackay (LM)), computed tomography (CT; LM) imaging, and sinonasal health-related quality of life (HRQOL) survey responses (SinoNasal Outcomes Test 22 (SNOT-22), Rhinosinusitis Disability Index (RSDI), and Patient Health Questionnaire 2 (PHQ2)) were obtained at enrollment. The SNOT-22 is a validated 22-item survey developed to evaluate symptom severity in CRS.9 Individual item scores are measured using patient selected responses on a Likert scale, where higher scores indicate worse symptom severity and HRQOL (score range 0–110). The RSDI is a 30-item survey instrument (comprised of 3 subdomains) that assesses the impact of rhinosinusitis on patient physical (score range: 0–44), functional (score range: 0–36), and emotional (score range: 0–40) status.10 Similar to the SNOT-22, higher scores indicate a worse HRQOL and greater negative daily function. The PHQ2 is a two-item survey that screens for depressive symptoms within the past two weeks.11 The LM scoring system (range: 0–24) estimates opacification severity in the maxillary, ethmoidal, sphenoidal, ostiomeatal complex, and frontal sinus regions on computed tomography (CT) imaging.12 The paranasal sinuses were evaluated bilaterally using rigid, 30-degree endoscopes (SCB Xenon 175; Karl Storz, Tuttlingen, Germany). Endoscopic exams were staged by the enrolling physician using the bilateral LK scoring system (score range, 0 −20), which quantifies pathologic states within the paranasal sinuses including the severity of polyposis, discharge, edema, scarring, and crusting on a Likert scale.13 Higher scores on both staging systems reflect worse disease severity.
Peripheral Blood Collection and Preparation for Gene Expression Studies
One to three milliliters of peripheral blood were obtained from each enrolled participant and placed into a K2-EDTA collection tube (Becton Dickinson; Franklin Lakes, NJ) prior to surgery. An aliquot of blood (0.5 mL) was added to 1 mL of RNAlater (AppliedBiosystems/Life Technologies Corp., Carlsbad, CA) and stored at −80°C until use. Blood samples were thawed at 4°C, and the RNAlater was removed after centrifugation (8000 rpm at 4°C; Labnet International; Edison, NJ) and aspiration. Nucleic acid was extracted using a RiboPure Blood Kit (Invitrogen; Carlsbad, CA) and quantified using a NanoDrop 8000 and Qubit (Thermo Fisher Scientific; Pittsburgh, PA), according to manufacturer’s protocols.
Gene Expression Analysis
Multiplex gene expression analysis of mRNA extracted from peripheral blood was performed by the Molecular Diagnostics Core at the Huntsman Cancer Institute (University of Utah, Salt Lake City, UT). A total of 594 genes broadly associated with innate and adaptive immunity in atopy, autoimmune, and infectious diseases were analyzed using NanoString technology (Human Immunology v2 Panel; Seattle, WA). Briefly, mRNA sample concentrations were normalized (25–300 ng) and added to a mixture containing hybridization buffer (70 µL), reporter probes (8 µL), and capture probes (2 µL). The mixture components were hybridized after 16 hours at 65°C and subjected to mRNA transcript copy count quantification using the nCounter® MAX equipped with a Digital Analyzer. mRNA transcript copy numbers for each gene and sample were generated after applying automated filters for background subtraction, quality control (QC), and normalization using nSolver version 4.0 software.
QC consisted of performing a correlation analysis in log2 space between the known concentrations of positive controls built into the panel for each gene. Data that were flagged during QC were omitted. Normalization was performed using the geometric mean of the positive control and 15 internal reference genes: ATP-binding cassette sub-family F member 1 (ABCF1), Delta-aminolevulinate synthase 1 (ALAS1), (Elongation factor 1-gamma) EEF1G, Glucose-6-phosphate dehydrogenase (G6PD), Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), beta-glucuronidase (GUSB), Hypoxanthine Phosphoribosyltransferase 1 (HPRT1), Ornithine decarboxylase antizyme (OAZ1), polymerase (RNA) I polypeptide B (POLRIB), RNA Polymerase II Subunit A (POLR2A), Peptidylprolyl isomerase A (PPIA), 60S ribosomal protein L19 (RPL19), Succinate Dehydrogenase Complex Flavoprotein Subunit A (SDHA), TATA-Box Binding Protein (TBP), and Tubulin Beta Class I (TUBB).
Statistical Analysis
Total counts were used to characterize categorical variables, while mean (standard deviation [SD]) or median (interquartile range) calculations were used to summarize continuous variables. All variables were stratified by diagnostic group (CRS-A vs CRS). Gene expression ratios were calculated and reported for genes that were significantly and differentially expressed between these cohorts. Student’s t-tests were employed to compare continuous variables between groups, while Chi-squared tests for independence or Fisher’s exact tests were used for comparing categorical variables. The threshold for significance was set at p < 0.05. Statistical analyses were conducted using R statistical software (R Core Team, Vienna, Austria, Version 3.6.1, 2019).
Results
Demographics
Thirty-four patients with CRS-A (n=19) and CRS (n=15) were included in the final analysis. There was no significant difference in age, gender, and nasal polyposis between the two groups (Table 1). Similarly, there was no significant difference in comorbidities between the two groups except for a history of migraines, which was present exclusively in the CRS cohort (p=0.02).
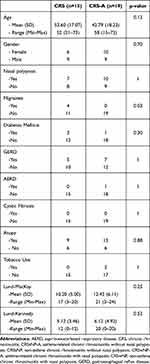 |
Table 1 Baseline Demographic Data for Patients with CRS Enrolled in This Study (N =34) |
SNOT-22
The mean baseline SNOT-22 score did not significantly differ between CRSsNP (57.8) and CRSsNP with asthma (CRSsNP-A) (50.5) (p=0.29). Similarly, the mean baseline SNOT 22 score did not significantly differ between CRSwNP (56.3) and CRSwNP with asthma (CRSwNP-A) (65) (p=0.52) (Table 2).
 |
Table 2 Patient-Reported Outcome Measure (PROM) Comparisons |
RSDI
Patients with CRSsNP had a significantly higher RSDI Functional subdomain score (18.5) compared to patients with CRSsNP-A (10) (p=0.03) (Table 2). However, there was no significant difference between the two groups in regard to the total RSDI score or the RSDI physical and emotional subdomain (Table 2). Similarly, there was no significant difference in any of the RSDI subdomains or the total RSDI score when comparing CRSwNP to CRSwNP-A (Table 2).
PHQ-2
The mean PHQ-2 scores were higher for both CRSwNP (2.6) and CRSwNP-A (2.4) compared to their CRSsNP counterparts (1.33 and 1.38), but there was no significant difference between the cohorts (Table 2).
Systemic Inflammation
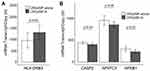
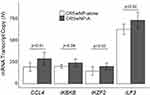
Of the 594 genes surveyed in peripheral blood, only HLA class II histocompatibility antigen, DRB3-1 beta chain (HLA-DRB3) was noted to be significantly upregulated in patients with CRSsNP-A compared to CRSsNP (p=0.04) (Table 3 and Figure 1A). Conversely, Caspase 2 (CASP2) (p=0.02), Nuclear factor of activated T-cells, cytoplasmic 3 (NFATC3) (p=0.04), and Nuclear factor NF-kappa-B p105 subunit (NFKB1) (p=0.02) were all noted to be significantly downregulated in this cohort compared to CRSsNP (Table 3 and Figure 1B). When examining the same genes in patients with CRSwNP, the following genes were noted to be upregulated in the asthma cohort compared to the non-asthma cohort: Chemokine (C-C motif) ligands 4 (CCL4) (p=0.01), inhibitor of nuclear factor kappa-B kinase subunit beta (IKBKB) (p=0.04), Zinc finger protein Helios (IKZF2) (p=0.02), and Interleukin enhancer-binding factor 3 (ILF3) (p=0.02) (Table 4 and Figure 2). A larger group of genes were noted to be significantly downregulated in the asthma cohort compared to the non-asthma cohort (p<0.04, Table 4 and Figure 3).
 |
Table 3 Inflammatory Gene Expression Ratio Comparisons in the Peripheral Blood of Patients with CRSsNP-A vs CRSsNP |
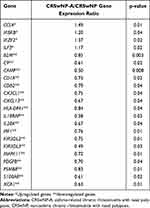 |
Table 4 Inflammatory Gene Expression in the Peripheral Blood of Patients with CRSwNP-A vs CRSwNP |
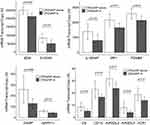 |
Figure 3 Downregulation of genes predominantly associated with Th1 and Th17 inflammation in peripheral blood favor an overall type 2 profile in patients with CRSwNP-A compared to CRSwNP. |
Discussion
Chronic rhinosinusitis with comorbid asthma presents a large healthcare burden and is associated with a significant patient morbidity.3 Despite this, limited research has investigated the underlying molecular endotypes within this clinical cohort. Herein, we demonstrated that systemic (ie, peripheral blood) genetic transcriptional changes in patients with CRS and asthma resulted in molecular endotypes associated with a type 2 inflammatory profile regardless of CRS phenotype.14–24 Additionally, although all patients with CRSwNP exhibited upregulation of genes associated with type 2 inflammation, this upregulation was the greatest for patients with concomitant asthma.
Type 2 inflammation is characterized by the presence of Th2 cells, whereas Type 1 and type 3 inflammation is characterized by Th1 and Th17 cells, respectively.7 Genetic transcriptional studies in asthmatics without CRS have demonstrated overrepresentation of genes associated with type 2 inflammation. Several key loci, including HLA polymorphisms14–16 are significantly upregulated in patients with asthma compared to healthy controls. HLA gene complex encodes major histocompatibility complex (MHC) class II molecule – a known integral component of the immune system.25 Polymorphisms near the class II HLA-DR alpha locus have also been implicated in immune disease and in the development of nasal polyposis.18,25 In the present study, we found significant upregulation of HLA-DRB3 in the peripheral blood of patients with CRSsNP-A compared to CRSsNP. Although class II HLA SNPs have previously been linked to CRS in the setting of nasal polyposis, to the best of our knowledge, this is the first study to implicate them in CRS, independent of polyp status. It appears that HLA polymorphisms may be uniquely upregulated in patients with asthma and remain so in the presence of comorbid CRS.
The upregulation of genes associated with type 2 inflammation was also observed in patients with CRSwNP, and more robustly for patients with concomitant asthma (ie, CRSwNP-A). IKBKB, an upstream regulator of NF-κB, and ILF3, a regulator of IL13 transcription were both upregulated in CRSwNP-A compared to CRSwNP.20,26 IKBKB has been shown to be upregulated in patients with asthma,16,20 and prior CRS data has shown a similar overrepresentation of type 2 inflammation in CRSwNP-A compared to CRSwNP.4,18,19,25 At the same time, in the present cohort, upregulation of these genes associated with type 2 inflammation was complemented by a simultaneous downregulation of genes associated with type 1 and type 3 inflammation. For example, compared to CRSwNP, patients with CRSwNP-A demonstrated downregulation of genes associated with Th17 differentiation (MAPK11)27 and Th1 signaling (PSMB8 and B2M).28,29 The upregulation of genes associated with type 2 inflammation with simultaneous downregulation of genes associated with type 1 and type 3 inflammation creates a more robust type 2 molecular endotype in CRSwNP-A compared to CRSwNP.
It is prudent to note that not all downstream mediators of type 2 inflammation were upregulated in the CRS-A cohort. Certain genes known to mediate Th2 inflammation in patients with asthma, such as the NFAT family of transcription factors,30 CASP2, and NF-κB, were in fact downregulated.16,23 It is possible that given the wide variability in the possible endotype and genetic profile of both asthma and CRS, many permutations of individual molecular endotypes are likely to be expressed. Some genes known to be overexpressed in asthma may dominate in some patients, while other genes known to be overexpressed in CRSsNP or CRSwNP may dominate in other patients. Unfortunately, we did not possess the data to endotype each patient’s asthma in this way, nor did we have a pure asthma cohort for comparison. Ultimately, the magnitude of upregulation of particular Th2 mediators, such as HLA-DRB3, as determined by gene copy numbers, was greater than the concomitant downregulation of other Th2 mediators (ie, NFAT, CASP2, and NF-κB) in CRSsNP-A.
CRS and asthma have tremendous impact on quality of life and treatment outcomes;2,31 this may lead to an expectation that CRS with comorbid asthma should have even worse overall HRQOL. However, our data demonstrated no significant difference between the asthma and non-asthma cohorts in regard to baseline SNOT-22, total RSDI, or PHQ-2 scores. This is consistent with prior work that has demonstrated no significant difference in baseline SNOT 22,32,33 RSDI,33 and PHQ-2 scores34 in CRS patients with and without asthma. Future work is required to further elucidate whether the dysregulation of particular genes can independently impact PROMs.
There are several limitations to consider while interpreting these findings, including sample size. Based on the criteria for performing genome-wide association studies, however, we believe the sample size to be ample.24 We chose to examine a select group of genes known to be associated with innate and adaptive immunity; thus, there may be other genetic transcriptional alterations among genes that were not examined. Further, significant changes in gene transcription do not always correlate to translation into a functional protein. Future research will include proteomic studies to corroborate our gene transcriptional findings. Despite these limitations, further defining molecular endotypes in patients with CRS with comorbid asthma provides a unique contribution to our field and serves as a key investigation for further mechanism-driven research.
This is the first investigation to characterize gene transcriptional changes in CRSsNP-A and correlate them with similar known genetic alterations in patients with asthma. Patients with CRSsNP-A demonstrated a molecular endotype associated with a robust type 2 inflammation compared to CRSsNP. Furthermore, patients with CRSwNP-A demonstrated increased overrepresentation of a type 2 molecular endotype compared to patients with CRSwNP, with a concomitant downregulation of genes associated with type 1 and type 3 inflammatory profile. Thus, CRS-A appears to lead to an upregulation of Th2 inflammation compared to CRS without asthma, independent of polyp phenotype.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
Abigail Pulsipher: Financial interests in GlycoMira Therapeutics.
Jeremiah A Alt: Financial interest and/or other relationships with GlycoMira Therapeutics, Inc. (Salt Lake City, UT; USA) is a consultant for Medtronic ENT and OptiNose, which are not affiliated with this research.
References
1. Centers for Disease Control and Prevention. National vital statistics reports. 2013;61(4).
2. Orlandi RR, Kingdom TT, Hwang PH, et al. International Consensus Statement on Allergy and Rhinology: rhinosinusitis. Int Forum Allergy Rhinol. 2016;6(Suppl 1):S22–209. doi:10.1002/alr.21695
3. Castagnoli R, Licari A, Brambilla I, Tosca M, Ciprandi G, Marseglia GL. An update on the role of chronic rhinosinusitis with nasal polyps as a co-morbidity in severe asthma. Expert Rev Respir Med. 2020:1–9.
4. Wang M, Bu X, Luan G, et al. Distinct type 2-high inflammation associated molecular signatures of chronic rhinosinusitis with nasal polyps with comorbid asthma. Clin Transl Allergy. 2020;10(1):26. doi:10.1186/s13601-020-00332-z
5. Stevens WW, Peters AT, Hirsch AG, et al. Clinical characteristics of patients with chronic rhinosinusitis with nasal polyps, asthma, and aspirin-exacerbated respiratory disease. J Allergy Clin Immunol Pract. 2017;5(4):1061–1070 e1063. doi:10.1016/j.jaip.2016.12.027
6. Smith KA, Orlandi RR, Oakley G, Meeks H, Curtin K, Alt JA. Long-term revision rates for endoscopic sinus surgery. Int Forum Allergy Rhinol. 2019;9(4):402–408. doi:10.1002/alr.22264
7. Ahern S, Cervin A. Inflammation and endotyping in chronic rhinosinusitis-a paradigm shift. Medicina (Kaunas). 2019;55(4). doi:10.3390/medicina55040095
8. Rosenfeld RM, Piccirillo JF, Chandrasekhar SS, et al. Clinical practice guideline (update): adult sinusitis. Otolaryngol Head Neck Surg. 2015;152:S1–S39. doi:10.1177/0194599815572097
9. Hopkins C, Gillett S, Slack R, Lund VJ, Browne JP. Psychometric validity of the 22-item Sinonasal Outcome Test. Clin Otolaryngol. 2009;34(5):447–454. doi:10.1111/j.1749-4486.2009.01995.x
10. Benninger MS, Senior BA. The development of the Rhinosinusitis Disability Index. Arch Otolaryngol Head Neck Surg. 1997;123(11):1175–1179. doi:10.1001/archotol.1997.01900110025004
11. Maurer DM, Raymond TJ, Davis BN. Depression: screening and diagnosis. Am Fam Physician. 2018;98(8):508–515.
12. Lund VJ, Mackay IS. Staging in rhinosinusitus. Rhinology. 1993;31(4):183–184.
13. Lund VJ, Kennedy DW. Staging for rhinosinusitis. Otolaryngol Head Neck Surg. 1997;117(3):S35–40. doi:10.1016/S0194-5998(97)70005-6
14. Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30.
15. Hsu J, Avila PC, Kern RC, Hayes MG, Schleimer RP, Pinto JM. Genetics of chronic rhinosinusitis: state of the field and directions forward. J Allergy Clin Immunol. 2013;131:
16. Seumois G, Zapardiel-Gonzalo J, White B, et al. Transcriptional profiling of Th2 cells identifies pathogenic features associated with asthma. J Immunol. 2016;197(2):655–664. doi:10.4049/jimmunol.1600397
17. Kim JY, Kim DK, Yu MS, Cha MJ, Yu SL, Kang J. Role of epigenetics in the pathogenesis of chronic rhinosinusitis with nasal polyps. Mol Med Rep. 2018;17:1219–1227. doi:10.3892/mmr.2017.8001
18. Bohman A, Juodakis J, Oscarsson M, Bacelis J, Bende M, Torinsson Naluai A. A family-based genome-wide association study of chronic rhinosinusitis with nasal polyps implicates several genes in the disease pathogenesis. PLoS One. 2017;12(12):e0185244. doi:10.1371/journal.pone.0185244
19. Plager DA, Kahl JC, Asmann YW, et al. Gene transcription changes in asthmatic chronic rhinosinusitis with nasal polyps and comparison to those in atopic dermatitis. PLoS One. 2010;5(7):e11450. doi:10.1371/journal.pone.0011450
20. Catley MC, Chivers JE, Holden NS, Barnes PJ, Newton R. Validation of IKK beta as therapeutic target in airway inflammatory disease by adenoviral-mediated delivery of dominant-negative IKK beta to pulmonary epithelial cells. Br J Pharmacol. 2005;145:114–122. doi:10.1038/sj.bjp.0706170
21. Tsai YH, Parker JS, Yang IV, Kelada SNP. Meta-analysis of airway epithelium gene expression in asthma. Eur Respir J. 2018;51(5):1701962. doi:10.1183/13993003.01962-2017
22. Shin SW, Oh TJ, Park SM, et al. Asthma-predictive genetic markers in gene expression profiling of peripheral blood mononuclear cells. Allergy Asthma Immunol Res. 2011;3(4):265–272. doi:10.4168/aair.2011.3.4.265
23. Janssen-Heininger YM, Poynter ME, Aesif SW, et al. Nuclear factor kappaB, airway epithelium, and asthma: avenues for redox control. Proc Am Thorac Soc. 2009;6(3):249–255. doi:10.1513/pats.200806-054RM
24. Liu M, Guo P, An J, Guo C, Lu F, Lei Y. Genomewide profiling of lncRNA and mRNA expression in CRSwNP. Mol Med Rep. 2019;19:3855–3863. doi:10.3892/mmr.2019.10005
25. Kim JH, Park BL, Cheong HS, et al. HLA-DRA polymorphisms associated with risk of nasal polyposis in asthmatic patients. Am J Rhinol Allergy. 2012;26:12–17. doi:10.2500/ajra.2012.26.3692
26. Kiesler P, Haynes PA, Shi L, Kao PN, Wysocki VH, Vercelli D. NF45 and NF90 regulate HS4-dependent interleukin-13 transcription in T cells. J Biol Chem. 2010;285(11):8256–8267. doi:10.1074/jbc.M109.041004
27. Huang G, Wang Y, Vogel P, Kanneganti TD, Otsu K, Chi H. Signaling via the kinase p38alpha programs dendritic cells to drive TH17 differentiation and autoimmune inflammation. Nat Immunol. 2012;13:152–161. doi:10.1038/ni.2207
28. Jones AC, Bosco A. Using network analysis to understand severe asthma phenotypes. Am J Respir Crit Care Med. 2017;195(11):1409–1411. doi:10.1164/rccm.201612-2572ED
29. Snahnicanova Z, Kasubova I, Kalman M, et al. Genetic and epigenetic analysis of the beta-2-microglobulin gene in microsatellite instable colorectal cancer. Clin Exp Med. 2020;20(1):87–95. doi:10.1007/s10238-019-00601-7
30. Koch S, Graser A, Mirzakhani H, et al. Increased expression of nuclear factor of activated T cells 1 drives IL-9-mediated allergic asthma. J Allergy Clin Immunol. 2016;137:1898–1902 e1897. doi:10.1016/j.jaci.2015.11.047
31. Stanescu S, Kirby SE, Thomas M, Yardley L, Ainsworth B. A systematic review of psychological, physical health factors, and quality of life in adult asthma. NPJ Prim Care Respir Med. 2019;29(1):37. doi:10.1038/s41533-019-0149-3
32. Penezic A, Paic M, Greguric T, Grgic MV, Baudoin T, Kalogjera L. The impact of asthma on quality of life and symptoms in patients with chronic rhinosinusitis. Curr Med Res Opin. 2020;36(6):1043–1048. doi:10.1080/03007995.2020.1754189
33. Quintanilla-Dieck L, Litvack JR, Mace JC, Smith TL. Comparison of disease-specific quality-of-life instruments in the assessment of chronic rhinosinusitis. Int Forum Allergy Rhinol. 2012;2(6):437–443. doi:10.1002/alr.21057
34. Schlosser RJ, Storck K, Cortese BM, Uhde TW, Rudmik L, Soler ZM. Depression in chronic rhinosinusitis: a controlled cohort study. Am J Rhinol Allergy. 2016;30(2):128–133. doi:10.2500/ajra.2016.30.4290
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.