Back to Journals » International Journal of Nanomedicine » Volume 12
The use of functional epirubicin liposomes to induce programmed death in refractory breast cancer
Authors Liu L, Mu LM, Yan Y, Wu JS, Hu YJ, Bu YZ, Zhang JY, Liu R, Li XQ, Lu WL
Received 25 January 2017
Accepted for publication 18 April 2017
Published 1 June 2017 Volume 2017:12 Pages 4163—4176
DOI https://doi.org/10.2147/IJN.S133194
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Linlin Sun
Lei Liu, Li-Min Mu, Yan Yan, Jia-Shuan Wu, Ying-Jie Hu, Ying-Zi Bu, Jing-Ying Zhang, Rui Liu, Xue-Qi Li, Wan-Liang Lu
Beijing Key Laboratory of Molecular Pharmaceutics and New Drug System, State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, Beijing, People’s Republic of China
Abstract: Currently, chemotherapy is less efficient in controlling the continued development of breast cancer because it cannot eliminate extrinsic and intrinsic refractory cancers. In this study, mitochondria were modified by functional epirubicin liposomes to eliminate refractory cancers through initiation of an apoptosis cascade. The efficacy and mechanism of epirubicin liposomes were investigated on human breast cancer cells in vitro and in vivo using flow cytometry, confocal microscopy, high-content screening system, in vivo imaging system, and tumor inhibition in mice. Mechanistic studies revealed that the liposomes could target the mitochondria, activate the apoptotic enzymes caspase 8, 9, and 3, upregulate the proapoptotic protein Bax while downregulating the antiapoptotic protein Mcl-1, and induce the generation of reactive oxygen species (ROS) through an apoptosis cascade. In xenografted mice bearing breast cancer, the epirubicin liposomes demonstrated prolonged blood circulation, significantly increased accumulation in tumor tissue, and robust anticancer efficacy. This study demonstrated that functional epirubicin liposomes could significantly induce programmed death of refractory breast cancer by activating caspases and ROS-related apoptotic signaling pathways, in addition to the direct killing effect of the anticancer drug itself. Thus, we present a simple nanomedicine strategy to treat refractory breast cancer.
Keywords: mitochondria, drug delivery, apoptosis, efficacy, signaling pathway
Introduction
Refractory breast cancer is responsible for the increasing mortality in women suffering from cancer.1 The current clinical treatment strategy is a comprehensive approach including surgery, radiotherapy, chemotherapy, and immunotherapy.2–4 Among these treatments, chemotherapy is crucial to eliminate cancer cells. However, it is far from satisfactory because of the existence of extensive multidrug resistance (MDR), resulting in a poor 5-year survival rate.5
MDR, which comprises both extrinsic and intrinsic drug resistance, represents a major obstacle to successful therapy.6 Extrinsic drug resistance derives from drug stimulus after administration, causing overexpression of ATP-binding cassette (ABC) transporter proteins on the cancer cell membrane, thereby leading to the efflux of the anticancer drug ingested by cancer cells.7,8 Intrinsic drug resistance is a naturally occurring drug tolerance in cancer cells, which could be associated with drug resistance of apoptosis-related genes or proteins of cancer cells.9,10 Cancer cells can be killed by anticancer agents via direct toxic injury and/or induced apoptosis. Therefore, apoptosis-related intrinsic drug resistance represents a complex problem.11
In this study, we hypothesize that functional epirubicin liposomes could be used for eliminating both extrinsic and intrinsic resistant breast cancer. In the liposomes, quinine was incorporated into the vesicles as a modulating agent, and dequalinium was incorporated into the bilayers as a mitochondrial targeting molecule.
Epirubicin is used as an antibiotic anticancer agent, which is widely employed to treat solid tumors, including breast cancer. Epirubicin is able to suppress DNA and RNA synthesis in cancer cells by directly embedding into base pairs of DNA to interfere with transcription.12 In addition, it suppresses the activity of topoisomerase II and upregulates the level of reactive oxygen species (ROS), and therefore kills cancer cells through varying mechanisms.13,14
Quinine is an alkaloid derived from the bark of the cinchona tree, and can be synthesized chemically as well. It is extensively applied in the treatment of malaria because of its antipyretic, analgesic, and anti-inflammatory properties. Our previous results showed that antimalarial agents such as quinacrine were useful agents for improving anticancer efficacy through blocking drug efflux and inducing apoptosis of refractory cancer or cancer stem cells.15,16 Accordingly, quinine was used as a potential modulating agent for eliminating resistant breast cancer cells in this study.
Dequalinium is an amphipathic cationic compound, and can selectively target the mitochondria of cancer cells driven by mitochondrial membrane negative potential.17 Therefore, it was used as a mitochondria-targeting molecule for improving the internalization of drug-loaded liposomes in cancer cells.18,19
Polyethyleneglycol-containing lipid nanovesicles have shown promising functions in circumventing refractory cancers. Increasing evidence has demonstrated that functionalized liposomes are able to maintain the nature of pegylated liposomes – which show enhanced permeability and retention (EPR) effects in tumor tissues – and to potentiate the killing effect in drug-resistant cancer and in cancer stem cells through varying mechanisms and signaling pathways.20–22
In the present study, we prepared functional epirubicin liposomes modified with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (DSPE-PEG2000) and dequalinium. The present study aimed to characterize the capability of these functional epirubicin liposomes to modulate mitochondria to eliminate refractory cancers.
Materials and methods
Materials and cells
Epirubicin hydrochloride was obtained from Nanjing Tianzun Zezhong Chemicals Co. Ltd. (Nanjing, People’s Republic of China). Quinine sulfate was purchased from Alfa Aesar Chemicals Co. Ltd. (Tianjin, People’s Republic of China). Dequalinium was purchased from Sigma-Aldrich Corporation (St Louis, MO, USA). DSPE-PEG2000 was purchased from Avanti Polar Lipids Inc. (Alabaster, AL, USA). SYTOX Green and Annexin V-keyfluor 647 were purchased from Keygen Biotech Co. Ltd. (Nanjing, People’s Republic of China). Other reagents were commercially available, and used without further purification.
Human breast cancer MCF-7 cells were obtained from the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, People’s Republic of China). Drug-resistant human breast cancer MCF-7/adr cells were obtained from the Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Sciences (Tianjin, People’s Republic of China). MCF-7 and MCF-7/adr cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Macgene, Beijing, People’s Republic of China) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA). Both cell lines were maintained in a 37°C humidified incubator with a 5% CO2 atmosphere. All cells in the experiments were in exponential growth phase, and no mycoplasmas were detected. Female BALB/c nude mice were purchased from the Peking University Experimental Animal Center (Beijing, People’s Republic of China).
Preparation of liposomes
Briefly, a mixture of egg phosphatidylcholine (EPC), cholesterol, DSPE-PEG2000, and dequalinium (55:25:3.5:8, molar ratio) was dissolved in chloroform/methanol (1:1, v/v). The solvent was evaporated using a rotary evaporator, and the lipid film was hydrated with 250 mM ammonium sulfate by gentle sonication in a water bath. The suspensions were sonicated using a probe-type sonicator for 10 min, followed by extrusion three times through 200 nm polycarbonate membranes (Millipore, Bedford, MA, USA). Thereafter, the blank functional liposomes were dialyzed against HEPES-buffered saline (HBS; 150 mM NaCl, 25 mM HEPES, pH 7.4) for 24 h. Drug loading was performed by adding the appropriate amount of epirubicin hydrochloride and quinine sulfate (1:1, molar ratio; lipids:drug =10:1, w/w). Subsequently, the liposomes were shaken intermittently for 30 min at 60°C in a water bath, yielding the functional epirubicin liposomes.
Epirubicin liposomes (encapsulating epirubicin only) and combinational epirubicin liposomes (encapsulating both epirubicin and quinine) were prepared using the same procedures as mentioned earlier, except for the addition of dequalinium. For in vivo imaging, functional dioctadecyltetramethyl indotricarbocyanine iodide (DiR) liposomes (lipids:DiR =300:1, w/w) were also prepared as controls using similar procedures.
Characterization of liposomes
The size distribution and zeta potential of the liposomes were measured using dynamic light scattering in a Nano Series Zen 4003 Zetasizer (Malvern Instruments Ltd., Malvern, UK). The morphology of the liposomes was observed using atomic force microscopy (SPI3800N series SPA-400; NSK Ltd., Tokyo, Japan).
Epirubicin, quinine, and dequalinium contents were measured using an octadecylsilyl groups column (Phenomenex, Torrance, CA, USA; 250×4.6 mm, 5 μm) in a high-performance liquid chromatography (HPLC) system with a ultraviolet (UV) detector (Shimadzu, Kyoto, Japan). The mobile phase to measure quinine comprised H2O (0.6% triethylamine) and acetonitrile (795:205, v/v, pH 3.5), and the detection wavelength was set at 275 nm. The mobile phase to measure epirubicin and dequalinium comprised acetonitrile, 0.02 M NaH2PO4, and triethylamine (34:66:0.3, v/v, pH 4.0), and the detection wavelength was set at 240 nm. The HPLC method was validated after evaluation of the linearity, recovery, detection limit, and interday and intraday precision.
The encapsulated and unencapsulated ingredients were separated by elution against a Sephadex G-50 column with HBS (pH 7.4). The entrapment efficiency (EE%) of epirubicin or quinine was calculated using the following equation: EE% = (Wencapsulated/Wtotal) ×100%, where Wencapsulated and Wtotal represent the amount of epirubicin or quinine in liposomes and the total amount of epirubicin or quinine, respectively. The modifying efficiency of dequalinium was determined using a similar method.
The in vitro release rates of epirubicin and quinine from the liposomes were determined by dialysis against phosphate-buffered saline (PBS; 137 mM NaCl, 2. 7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4) containing 10% FBS. Epirubicin and quinine were then quantified using HPLC as described earlier.
Cytotoxicity
The cytotoxic effects of functional epirubicin liposomes and other drug formulations against human breast cancer MCF-7 or MCF-7/adr cells (drug-refractory mutant) were determined using the sulforhodamine B assay. The absorbance of cells in each well was measured at 540 nm using a microplate reader (Tecan Infinite F50; Tecan Group Ltd., Männedorf, Switzerland). SPSS 20.0 software was used to calculate the IC50 value (the concentration of an inhibitor where the response [or binding] is reduced by half).
Blocking drug efflux
MCF-7/adr cells were seeded in six-well plates at a density of approximately 3×105 cells/well for 24 h. After 12 h of incubation with functional epirubicin liposomes or the other drug formulations, cells were harvested, resuspended in ice-cold PBS (pH 7.4), and subjected immediately to flow cytometry (Guava; EMD Millipore).
Colocalization into mitochondria
The MCF-7/adr cells were grown to 40%–50% confluence in chambered coverslips. After 6 h of incubation with functional epirubicin liposomes or the other drug formulations, these cells were stained with Mitotracker Deep Red and Hoechst 33342. The coverslips were then visualized under a confocal laser scanning microscope (Leica, Wetzlar, Germany). The velocity software was used to analyze the raw data and calculate the colocalization coefficients.
Apoptosis induction
The MCF-7/adr cells were seeded in six-well plates for 24 h, and incubated with functional epirubicin liposomes or the other drug formulations. After 24 h, the cells were stained with SYTOX Green and Annexin V-keyfluor 647, according to the manufacturer’s protocol. Once stained, the cells were subjected to flow cytometric detection for 1 h (Gallios, Beckman Coulter, Brea, CA, USA).
Apoptotic morphology
The MCF-7/adr cells were seeded in 12-well plates for 24 h, and incubated with various epirubicin formulations. After 12 h, these cells were stained with Mitotracker Deep Red and Hoechst 33342, and observed using the Operetta high-content screening system (PerkinElmer, Waltham, MA, USA).
Apoptosis signaling pathways
The MCF-7/adr cells were grown to 40%–50% confluence in 96-well plates, and incubated with various epirubicin formulations. After 6 h, the cells were fixed, permeabilized, blocked, and finally incubated with primary antibodies (anti-caspase 3, anti-caspase 8, anti-caspase 9, anti-Mcl-1 [Sangon, Shanghai, People’s Republic of China], anti-Bax, and anti-cytochrome C [Beyotime, Shanghai, People’s Republic of China]). The cells were detected after staining with Alexa fluor-488-labeled secondary antibody (OriGene, Rockville, MD, USA) and Hoechst 33342. The fluorescence intensity of each well was measured using the Operetta high-content screening system (PerkinElmer).
The MCF-7/adr cells were seeded in six-well plates at a density of approximately 3×105 cells/well in 1.8 mL of complete RPMI-1640 medium for 24 h. Free drugs (0.2 mL) and different liposome formulations were added to make a final epirubicin concentration of 20 μM. Free RPMI-1640 medium was added as a blank control. After 12 h of incubation, the cells were stained with 1 μM dichloro-dihydro-fluorescein diacetate (DCFH-DA) for 10 min. The cells were then washed, harvested, resuspended in ice-cold PBS, and subjected immediately to flow cytometric detection (Guava).
Anticancer efficacy and bio-distribution
All in vivo experimental procedures were conducted according to the guidelines of the Institutional Authority for Laboratory Animal Care of Peking University and were conducted with approval from the Laboratory Animal Welfare and Ethical Sub-committee at the Bioethics Committee of Peking University. Briefly, BALB/c female nude mice (6 weeks old) were inoculated subcutaneously with 1×107 MCF-7/adr cells. Tumors were allowed to grow to a volume of approximately 1,000 mm3 before extraction. Extracted tumors were then seeded into fresh nude mice. After 13 days, the mice were injected intravenously through the tail vein with physiological saline, epirubicin (5 mg/kg), epirubicin liposomes (5 mg/kg epirubicin), antiresistant epirubicin plus quinine liposomes (5 mg/kg epirubicin), or antiresistant targeting epirubicin plus quinine liposomes (5 mg/kg epirubicin) every other day (six mice per group). Tumor sizes were measured using Vernier calipers every day after drug administration, and the tumor volumes were calculated by the formula: V=0.5× length × width2. The inhibitory ratio of tumor volume was calculated using the following formula: the inhibitory ratio of tumor volume (%) at day n=100× Vn/V13, where Vn is the tumor volume at day n, and V13 is the tumor volume at day 13.
BALB/c female nude mice (6 weeks old) were inoculated subcutaneously with 1×107 MCF-7/adr cells. When the tumors reached approximately 500 mm3, the mice were administered physiological saline, free DiR (2 mg/kg), and targeted DiR liposomes (2 mg/kg) via tail-vein injection. Mice were scanned at 1, 3, 6, 12, 24, and 36 h using a Kodak multimode imaging system (Carestream Health, Inc., Rochester, NY, USA) after being anesthetized by isoflurane. For each mouse, at least three fields were observed, and one field was captured. The mice were sacrificed, and their major organs were extracted at 36 h after injection. Fluorescence signal intensities in the different organs were measured.
Statistical analysis
Data were expressed as the mean ± standard deviation, and statistical significance was assessed by analysis of variance with a Bonferroni post hoc test (P<0.05).
Results
Characterization of the liposomes
Table 1 shows the results for characterization of the prepared liposomes. The mean particle sizes of blank functional liposomes, epirubicin liposomes, combinational epirubicin liposomes, and functional epirubicin liposomes were in the range of 98.46±0.27 and 102.47±0.40 nm. Their zeta potential values were in the range of −6.63±0.11 and −8.28±0.41. The apparent slightly negative zeta potential values could provide a longer physical stability for liposome particles in the suspensions. The polydispersity index (PDI) for the liposomes was approximately 0.2, and the narrow PDI values represented the homogeneity of liposomes. In addition, for all liposomes, the encapsulation efficiency of epirubicin was ≥90% and that of quinine was ≥85%. In the functional epirubicin liposomes, the modification rate of the targeting molecule dequalinium was 85.5%±0.5%. The encapsulation efficiency represents the drug-loading efficiency, and a higher loading efficiency (>80%) could meet the requirement of potential industrial production by decreasing drug loss during preparation.
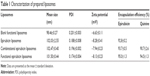  | Table 1 Characterization of prepared liposomes |
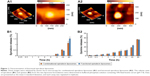
Figure 1A1 and A2 demonstrates atomic force microscopy images of one functional epirubicin liposome and one combinational epirubicin liposome. Both liposomes were round with smooth surfaces, and particle size was approximately 100 nm.
Figure 1B1 and B2 shows release rates of epirubicin and quinine from combinational epirubicin or functional epirubicin liposomes. At 48 h, the release rates of epirubicin from the combinational epirubicin liposomes and the functional epirubicin liposomes were 1.12%±0.16% and 1.51%±0.19%, respectively. Similarly, at 48 h, the release rates of quinine from combinational epirubicin and functional epirubicin liposomes were 36.80%±0.86% and 36.23%±1.32%, respectively. The release rate of dequalinium from the liposomes was negligible and below the detection threshold.
Cytotoxicity to breast cancer cells
Figure 2 displays the inhibitory effects on MCF-7 cells (Figure 2A and B) or MCF-7/adr cells (Figure 2C and D) after various treatments. MCF-7 cells were sensitive to free epirubicin alone and to free epirubicin plus quinine. Quinine alone did not show any cytotoxicity (Figure 2A). For liposome formulations, the ranking by antiproliferative effects against MCF-7 cells was functional epirubicin liposomes ≥ combinational epirubicin liposomes > epirubicin liposomes > blank functional liposomes (Figure 2B).
The MCF-7/adr cells were resistant to free epirubicin alone but sensitive to free epirubicin plus quinine, showing a quinine dose-dependent cytotoxic effect (Figure 2C). For the liposome formulations, the ranking by antiproliferative effects against MCF-7/adr cells was functional epirubicin liposomes > combinational epirubicin liposomes > epirubicin liposomes > blank functional liposomes (Figure 2D).
Table S1 shows the IC50 values of various drug formulations against MCF-7 and MCF-7/adr cells. The IC50 values of free epirubicin, free epirubicin plus quinine, epirubicin liposomes, combinational epirubicin liposomes, and functional epirubicin liposomes against MCF-7 cells were 0.48±0.05, 0.35±0.02, 0.83±0.09, 0.57±0.04, and 0.48±0.01 μM, respectively. The IC50 values of those against MCF-7/adr cells were 25.8±0.2, 2.5±0.2, 61.4±5.4, 15.4±1.0, and 6.5±0.7 μM, respectively. Resistance indices of these abovementioned formulations were 53.8, 7.1, 74, 27, and 13.5, respectively.
Blocking drug efflux from breast cancer cells
The ranking of cellular uptake by MCF-7/adr cells was free epirubicin plus quinine > functional epirubicin liposomes = combinational epirubicin liposomes > free epirubicin > epirubicin liposomes (Figure S1).
Colocalization into mitochondria of breast cancer cells
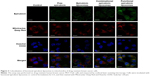
Figure 3 illustrates the images captured by confocal laser scanning microscopy of the colocalization of drug formulations in the mitochondria of MCF-7/adr cells. Mitochondria that were stained by Mitotracker Deep Red showed a red pseudocolor, whereas epirubicin manifested a green pseudocolor. Yellow spots in the merged pictures indicated the colocalization of epirubicin within mitochondria. The functional epirubicin liposomes accumulated selectively in the mitochondria of drug-resistant cancer cells.
In addition, green spots around cells in the merged pictures could indicate the efflux of epirubicin from drug-resistant cancer cells. These results intuitively exhibited that the efflux of epirubicin was the lowest after treatment with functional epirubicin liposomes, as compared to treatment with other formulations.
Inducing apoptosis of breast cancer cells
Figure 4 depicts the induced apoptosis of MCF-7/adr cells. After 24 h incubation with free epirubicin, free epirubicin plus quinine, epirubicin liposomes, combinational epirubicin liposomes, and functional epirubicin liposomes, the percentages of total apoptosis were 18.99%, 57.99%, 16.72%, 60.37%, and 70.28%, respectively.
Apoptotic morphology of breast cancer cells
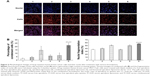
Figure 5A shows the fluorescence images captured by the high-content screening system for nuclei and whole MCF-7/adr cells after treatment with functional epirubicin liposomes. Nuclei were stained with Hoechst 33342, and mitochondria were stained with Mitotracker Deep Red to outline the cells. Consequently, bright blue spots indicated condensed nuclei, whereas bright red spots represented condensed cells. After treatment with functional epirubicin liposomes, more condensed nuclei and cells were observed, compared with cells treated with the other formulations. Meanwhile, the blue spots could be matched well with the red spots, suggesting that cell shrinkage was accompanied by nuclear condensation.
Figure 5B and C indicates the percentages of condensed cells and the fragmentation indices of nuclei, estimated using the Columbus system. After treatment with blank medium, free epirubicin, free epirubicin plus quinine, epirubicin liposomes, combinational epirubicin liposomes, and functional epirubicin liposomes, the percentages of condensed cells were 2.49%±1.00%, 4.29%±1.56%, 14.70%±5.79%, 6.24%±2.03%, 7.13%±3.55%, and 21.88%±3.60%, respectively (Figure 5B). The fragmentation indices of nuclei (the coefficient of variation of nuclei area) were 37.04%±1.46%, 40.57%±1.23%, 51.07%±2.06%, 36.66%±1.39%, 50.31%±2.78%, and 56.33%±2.25%, respectively (Figure 5C).
Apoptosis signaling pathways of breast cancer cells
Figure S2 displays the quantitative results of apoptotic enzymes and apoptotic proteins in MCF-7/adr cells after various treatments. After exposure to blank medium, epirubicin liposomes, combinational epirubicin liposomes, and functional epirubicin liposomes, the activity ratios of caspase 8 were 1.00±0.02, 1.00±0.03, 1.06±0.06, and 1.21±0.07 (Figure S2A); the activity ratios of Bax were 1.00±0.02, 1.04±0.03, 1.12±0.01, and 1.17±0.03 (Figure S2B); the activity ratios of Mcl-1 were 1.00±0.03, 1.02±0.06, 0.95±0.02, and 0.89±0.03 (Figure S2C); the activity ratios of cytochrome C were 1.00±0.03, 1.07±0.03, 1.10±0.02, and 1.17±0.04 (Figure S2D); the activity ratios of caspase 9 were 1.00±0.04, 1.04±0.05, 1.06±0.03, and 1.22±0.05 (Figure S2E); and the activity ratios of caspase 3 were 1.00±0.03, 1.03±0.03, 1.03±0.05, and 1.17±0.08 (Figure S2F), respectively.
Figure S3 shows the ROS levels in the MCF-7/adr cells after various treatments. The ranking by ROS levels was: functional epirubicin liposomes > combinational epirubicin liposomes = free epirubicin plus quinine > epirubicin liposomes > free epirubicin.
Anticancer efficacy and distribution in breast cancer-bearing mice
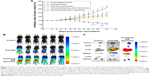
Figure 6A shows the tumor volume ratios over time in MCF-7/adr-cell-xenografted mice after various treatments. At day 24, the ranking by tumor volume ratios after administration was: physiological saline > epirubicin liposomes > free epirubicin > combinational epirubicin liposomes > functional epirubicin liposomes.
Figure 6B depicts the real-time distribution of the fluorescence marker DiR in the refractory breast cancer-bearing mice after intravenous injection with physiological saline, free DiR, and functional DiR liposomes. A strong fluorescence signal was observed in tumor-bearing mice after injection with functional DiR liposomes, and the signal was maintained at least up to 36 h. Furthermore, functional DiR liposomes accumulated obviously in tumor tissues. By contrast, after injection with free DiR, the fluorescence signal faded sharply after 3 h, and free DiR only accumulated minimally in tumor tissue and was distributed mainly in liver tissue.
Figure 6C further indicates the accumulation of DiR in the extracted organs at 36 h after the injection. The fluorescence signal was detected only in the liver and spleen after administration of free DiR. By contrast, bright fluorescence signals were observed in the tumor, heart, liver, spleen, and kidney tissues after administration of functional DiR liposomes, indicating that the functional DiR liposomes were retained in the circulation system for a longer time and showed higher accumulation in tumor tissues as compared to free DiR.
Discussion
Chemotherapy plays a key role in the comprehensive strategy to fight breast cancer. However, in recent decades, conventional chemotherapy using a single cytotoxic anticancer agent appears to be less efficient in controlling the continued development of the cancer in clinical treatment.23,24 Conventional chemotherapy is unable to eliminate cancer cells completely. An important springhead is multidrug resistance, which comprises extrinsic and intrinsic resistance. Functional drug liposomes may offer a new method to overcome these issues by accumulating more drugs in tumor tissues, inhibiting the efflux of anticancer drugs ingested by cancer cells, and initiating cancer cell death by targeting organelles and apoptosis-related genes or proteins.
Therefore, we developed functional epirubicin liposomes to overcome both types of drug resistance. Extrinsic drug resistance results from the overexpression of ABC transporters on cancer cell membranes, which causes efflux of the internalized anticancer drug. Accordingly, quinine was used as a regulating agent to block the efflux of epirubicin.25 Intrinsic drug resistance is associated with the inactivation of apoptosis-related genes or proteins, thereby blocking the initiation of apoptosis in cancer cells after regular chemotherapy. Therefore, dequalinium was used as a targeting molecule to deliver the drug-loaded liposome carriers onto the mitochondria of cancer cells. In addition to possessing superior functions of previously fabricated drug liposomes,26,27 such as prolonged circulation in blood by avoiding rapid clearance through the reticuloendothelial system (RES), the altered drug distribution thereby decreasing systemic toxicity (such as reduction of the cardiotoxicity of doxorubicin), and passively targeted accumulation in tumor tissue by the EPR effect, the functional epirubicin liposomes in this study were assigned new functions, including targeting mitochondria and blocking drug efflux, thereby being able to kill both extrinsically and intrinsically resistant cancer cells. Besides, the encapsulation procedure of functional epirubicin liposomes was an easy approach, and could be promising for future clinical use. Nevertheless, the main limitation for this kind of drug liposomes is that they might face more complicated evaluation procedures during new drug application due to combination use of two drugs.
Results from the in vitro cytotoxicity test demonstrated that quinine could enhance the cytotoxicity of epirubicin against MCF-7 and MCF-7/adr cells (Figure 2A and C). One of the mechanisms for this effect might be quinine-induced inhibition of epirubicin efflux (Figures S1 and 3).28 Both epirubicin and quinine are substrates of ABC transporters, and thus, additional quinine would occupy some of the transporters, allowing more epirubicin to be retained in the cancer cells. After addition of dequalinium to the functional liposomes, drug-loaded liposomes were colocalized in mitochondria via the interaction between the positive charges of dequalinium and the negative charges of mitochondria.29 Mitochondria are major organelles involved in cell apoptosis.30 Consequently, the mitochondrial targeting of epirubicin and quinine carried by the liposomes induced apoptosis through the pathway.
Apoptosis analysis by flow cytometry (Figure 4) and morphological assays using the high-content screening system (Figure 5) revealed that apoptosis occurred after treatment with functional epirubicin liposomes. In apoptotic cells, phosphatidyl serine is translocated from the inner to the outer leaflet of the cell membrane, resulting in increased binding of Annexin V.31 The integrity of the cell membrane is compromised in late apoptotic cells; thus, SYTOX Green could permeate and bind to nuclear DNA.32 Intact cancer cells possess the largest nuclei, whereas early apoptotic nuclei become condensed before they eventually become fragmented. In the late stage of apoptosis, shrunken cells appear along with the fragmented nuclei. In this study, MCF-7/adr cells treated with functional epirubicin liposomes exhibited the highest apoptotic percentages, the highest percentages of condensed cells, and the largest nuclear fragmentation index (Figures 4 and 5).
When functional epirubicin liposomes were internalized by cancer cells, two things happened: the liposomes ruptured or remained intact. Accordingly, the partial release or leakage of epirubicin and quinine into the cytoplasm during cancer cellular uptake caused nuclear attack, blocked efflux, generated ROS, and activated apoptotic enzymes, thus causing direct cytotoxic injury by interfering with transcription of DNA, and inducing programmed death of cancer cells by initiating apoptotic pathways.33 Meanwhile, intact targeting drugs-loaded liposomes were ingested by mitochondria through an electrical attraction followed by phagocytosis (Figure 2). The released drug in the mitochondria further caused the release of cytochrome C and a cascade of apoptotic reactions.34
Apoptotic mechanisms involve three aspects:35 (1) external signal (extrinsic or death receptor pathway), which activates initiator caspase 8, followed by the induction of the activation of the effector caspases (eg, caspase 3); (2) internal signals arising within the cell (the intrinsic or mitochondrial pathway), which activate initiator caspase 9, followed by the induction of the activation of the effector caspases (eg, caspase 3); and (3) apoptosis triggered by ROS, which does not use caspases. This defines an important profile for cell apoptotic pathways; however, the process is incompletely understood.
Upregulation of caspase 8 is indispensable for the complex signal transduction in the extrinsic apoptotic pathway (Figure S2A). Both proapoptotic protein (Bax) and antiapoptotic protein (Mcl-1) are located on the mitochondrial membrane. These apoptosis-related proteins maintain the balance between cell survival and death, and generally, they are involved in intrinsic drug resistance.36
After colocalization of functional epirubicin liposomes into mitochondria, the released epirubicin and quinine upregulated the proapoptotic protein Bax, but downregulated the antiapoptotic protein Mcl-1 (Figure S2B and C). The activation of Bax, the inhibition of Mcl-1, the release of cytochrome C (Figure S2D), and the subsequent activation of caspases 9 and 3 (Figure S2E and F) would result in a cascade of apoptotic reactions.37
Furthermore, recent studies revealed that the ROS mediate mitochondrial permeability transition, which results in increased expression of apoptotic proteins, such as Bax and caspase 9.38 The ROS thus generated might be involved in damaging the mitochondrial membrane, and might contribute partially to the release of cytochrome C, the upregulation of Bax, and the activation of caspase 9, thereby inducing cell apoptosis via the mitochondrial pathway.
The antitumor study in mice demonstrated the therapeutic efficacy of the functional epirubicin liposomes in treating breast cancer (Figure S3). This enhanced efficacy could have been mediated as follows: first, PEGylated liposomes evade rapid clearance by the RES in circulating blood; second, the appropriate size of liposomes results in increased accumulation in cancer tissue by EPR effect;39 third, quinine could inhibit the efflux of epirubicin; and finally, dequalinium could mediate liposomal targeting to the mitochondria to trigger apoptosis.
Conclusion
In the present study, functional epirubicin liposomes, with a size of approximately 100 nm, were developed successfully to treat drug-resistant breast cancer. They were able to enhance the uptake of epirubicin by breast cancer cells, caused direct acute cytotoxic injury to the cells, were targeted to the mitochondria, activated the apoptotic enzyme caspase 8, upregulated the proapoptotic protein Bax, downregulated the antiapoptotic protein Mcl-1, and increased ROS levels, followed by activation of caspase 9 and caspase 3, which initiated a cascade of apoptotic reactions. Functional epirubicin liposomes demonstrated strong efficacy against drug-resistant breast cancer in vitro and in mice. This study showed that the functional epirubicin liposomes could induce the programmed death of the refractory breast cancer significantly by activating caspases and ROS-related apoptotic signaling pathways, in addition to the direct killing effect of the anticancer drug, thus providing a simple nanomedicine strategy against refractory breast cancer.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program) (grant number 2013CB932501), the Beijing Natural Science Foundation (grant number 7131009), and the National Natural Science Foundation of China (grant number 81373343).
Disclosure
The authors report no conflicts of interest in this work.
References
Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1–22. | ||
Thariat J, Kirova Y, Milano G, Mornex F. Association d’une chimiothérapie ou d’un traitement ciblé à une irradiation stéréotaxique : état des lieux et recommandations préliminaires. [Combination of stereotactic irradiation and chemotherapy or targeted therapies: state of the art and preliminary recommendations]. Cancer Radiother. 2014;18(4):270–279. French [with English abstract]. | ||
Rekers NH, Troost EG, Zegers CM, Germeraad WT, Dubois LJ, Lambin P. Stereotactic ablative body radiotherapy combined with immunotherapy: present status and future perspectives. Cancer Radiother. 2014;18(5–6):391–395. | ||
Khong A, Cleaver AL, Fahmi Alatas M, et al. The efficacy of tumor debulking surgery is improved by adjuvant immunotherapy using imiquimod and anti-CD40. BMC Cancer. 2014;14:969. | ||
Katanoda K, Matsuda T. Five-year relative survival rate of breast cancer in the USA, Europe and Japan. Jpn J Clin Oncol. 2014;44(6):611. | ||
Moscow JA, Cowan KH. Multidrug resistance. Cancer Chemother Biol Response Modif. 1990;11:97–114. | ||
Saeki T, Tsuruo T, Sato W, Nishikawsa K. Drug resistance in chemotherapy for breast cancer. Cancer Chemother Pharmacol. 2005;56 Suppl 1:84–89. | ||
Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. | ||
Fojo AT, Shen DW, Mickley LA, Pastan I, Gottesman MM. Intrinsic drug resistance in human kidney cancer is associated with expression of a human multidrug-resistance gene. J Clin Oncol. 1987;5(12):1922–1927. | ||
Yang KP, Liang YF, Samaan NA. Intrinsic drug resistance in a human medullary thyroid carcinoma cell line: association with overexpression of mdrl gene and low proliferation fraction. Anticancer Res. 1991;11(3):1065–1068. | ||
Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–627. | ||
Plosker GL, Faulds D. Epirubicin: a review of its pharmacodynamic and pharmacokinetic properties, and therapeutic use in cancer chemotherapy. Drugs. 1993;45(5):788–856. | ||
Davies SL, Popert R, Coptcoat M, Hickson ID, Masters JR. Response to epirubicin in patients with superficial bladder cancer and expression of the topoisomerase II alpha and beta genes. Int J Cancer. 1996;65(1):63–66. | ||
Lo YL, Wang W. Formononetin potentiates epirubicin-induced apoptosis via ROS production in HeLa cells in vitro. Chem Biol Interact. 2013;205(3):188–197. | ||
Zhang L, Yao HJ, Yu Y, et al. Mitochondrial targeting liposomes incorporating daunorubicin and quinacrine for treatment of relapsed breast cancer arising from cancer stem cells. Biomaterials. 2012;33(2):565–582. | ||
Li XY, Zhao Y, Sun MG, et al. Multifunctional liposomes loaded with paclitaxel and artemether for treatment of invasive brain glioma. Biomaterials. 2014;35(21):5591–5604. | ||
Weissig V. Targeted drug delivery to mammalian mitochondria in living cells. Expert Opin Drug Deliv. 2005;2(1):89–102. | ||
D’Souza GG, Boddapati SV, Weissig V. Mitochondrial leader sequence – plasmid DNA conjugates delivered into mammalian cells by DQAsomes co-localize with mitochondria. Mitochondrion. 2005;5(5):352–358. | ||
Zhang Y, Li RJ, Ying X, et al. Targeting therapy with mitosomal daunorubicin plus amlodipine has the potential to circumvent intrinsic resistant breast cancer. Mol Pharm. 2011;8(1):162–175. | ||
Li RJ, Ying X, Zhang Y, et al. All-trans retinoic acid stealth liposomes prevent the relapse of breast cancer arising from the cancer stem cells. J Control Release. 2011;149(3):281–291. | ||
Mamot C, Drummond DC, Hong K, Kirpotin DB, Park JW. Liposome-based approaches to overcome anticancer drug resistance. Drug Resist Updat. 2003;6(5):271–279. | ||
Booser DJ, Esteva FJ, Rivera E, et al. Phase II study of liposomal annamycin in the treatment of doxorubicin-resistant breast cancer. Cancer Chemother Pharmacol. 2002;50(1):6–8. | ||
Calvo E. Systemic chemotherapy for metastatic colorectal cancer: reasons to combine. Clin Colorectal Cancer. 2002;2(3):170–172. | ||
Gonzalez-Angulo AM, Hortobagyi GN. Is there an ideal way to combine trastuzumab with chemotherapy? J Clin Oncol. 2011;29(34):4474–4476. | ||
Solary E, Velay I, Chauffert B, et al. Sufficient levels of quinine in the serum circumvent the multidrug resistance of the human leukemic cell line K562/ADM. Cancer. 1991;68(8):1714–1719. | ||
Xu Y, Qiu L. Nonspecifically enhanced therapeutic effects of vincristine on multidrug-resistant cancers when coencapsulated with quinine in liposomes. Int J Nanomedicine. 2015;10:4225–4237. | ||
Plourde K, Derbali RM, Desrosiers A, Dubath C, Vallée-Bélisle A, Leblond J. Aptamer-based liposomes improve specific drug loading and release. J Control Release. 2017;251:82–91. | ||
Morjani H, Belhoussine R, Lahlil R, Manfait M. Pirarubicin nuclear uptake does not correlate with its induced cell death effect during reversal of multidrug resistance by quinine in human K562 and CEM leukemic cells. Eur J Haematol. 1998;61(4):240–249. | ||
Weissig V, Torchilin VP. Mitochondriotropic cationic vesicles: a strategy towards mitochondrial gene therapy. Curr Pharm Biotechnol. 2000;1(4):325–346. | ||
Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407(6805):770–776. | ||
Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood. 1994;84(5):1415–1420. | ||
Roth BL, Poot M, Yue ST, Millard PJ. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl Environ Microbiol. 1997;63(6):2421–2431. | ||
Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5(11):897–907. | ||
Elkholi R, Renault TT, Serasinghe MN, Chipuk JE. Putting the pieces together: how is the mitochondrial pathway of apoptosis regulated in cancer and chemotherapy? Cancer Metab. 2014;2:16. | ||
Potten C, Wilson J. Apoptosis. Cambridge: Cambridge University Press; 2004. | ||
Green DR. Apoptotic pathways: paper wraps stone blunts scissors. Cell. 2000;102(1):1–4. | ||
Li X, Ruan GR, Lu WL, et al. A novel stealth liposomal topotecan with amlodipine: apoptotic effect is associated with deletion of intracellular Ca2+ by amlodipine thus leading to an enhanced antitumor activity in leukemia. J Control Release. 2006;112(2):186–198. | ||
Paul S, Sengupta S, Bandyopadhyay TK, Bhattacharyya A. Stevioside induced ROS-mediated apoptosis through mitochondrial pathway in human breast cancer cell line MCF-7. Nutr Cancer. 2012;64(7):1087–1094. | ||
Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46(12 Pt 1):6387–6392. |
Supplementary materials
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.