Back to Journals » OncoTargets and Therapy » Volume 11
The RNASEL −1385G/A polymorphism is associated with risk of prostate cancer in Africans
Authors Liu X, Zheng D, Lu G, Yang B
Received 11 September 2017
Accepted for publication 21 November 2017
Published 22 December 2017 Volume 2018:11 Pages 97—102
DOI https://doi.org/10.2147/OTT.S151398
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Xiaolei Liu,1 Dejie Zheng,2 Guowei Lu,3 Baohong Yang2
1Respiratory Department, Weifang Medical University, 2Department of Oncology, Weifang People’s Hospital, Weifang, 3Department of Otorhinolaryngology Head and Neck Surgery, The Affiliated Yantai Yuhuangding Hospital, Qingdao University, Qingdao, People’s Republic of China
Abstract: The RNASEL –1385G/A (rs486907) variant has been reported to be associated with increased risk of prostate cancer. However, these associations are not consistent among studies. To address this issue, we performed a meta-analysis to evaluate the association between RNASEL –1385G/A polymorphism and prostate cancer risk. The PubMed, Embase, and Web of Science databases were searched for relevant papers published in the past 20 years from 1997 to 2017. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the strength of associations. Based on our search for manuscripts reporting prostate cancer susceptibility related to the rs486907 polymorphism, 16 case–control studies from 13 different publications were retrieved. No significantly positive associations were found for the polymorphism and prostate cancer susceptibility in the total population. When stratified by ethnicity, the results demonstrated that the –1385G/A polymorphism was associated with a decreased cancer risk in Africans (GG vs AA: OR =0.371, 95% CI =0.176–0.783; GG/GA vs AA: OR =0.368, 95% CI =0.175–0.776). We also found that the rs486907 polymorphism was associated with a decreased cancer risk in hospital-based controls (GG vs AA: OR =0.697, 95% CI =0.488–0.996; GG + GA vs AA: OR =0.701, 95% CI =0.502–0.978). Our meta-analysis suggests that polymorphism in the RNASEL gene is a protective factor against prostate cancer in Africans. Further studies using larger sample sizes should be conducted to elucidate the role of gene polymorphism in prostate cancer risk.
Keywords: RNASEL, prostate cancer, polymorphism, meta-analysis
Introduction
RNase L, encoded by the RNASEL gene, is a ubiquitously expressed endoribonuclease.1 It is activated by dimerization, which occurs upon 2′–5′A binding, and results in cleavage of all RNAs in the cell. This can lead to activation of MDA5, an RNA helicase involved in the production of interferons.2 Beyond its antiviral activity, RNASEL contributes toward innate immunity and cell metabolism including inflammation, cell proliferation and differentiation, migration, apoptosis, and tumorigenesis.3–5
Several variations in RNASEL have been identified, of which the mutation at codon 462 is functionally distinct: the Arg-to-Gln variant has a threefold reduced enzyme activity, which could enhance virus susceptibility, impair the cellular stress response, diminish control of cellular RNA levels, or induce apoptosis.6
Over the years, the rs486907 G/A polymorphism in the RNASEL gene has been studied to assess its association with risk of prostate cancer.7,10–13 However, the results are inconsistent, which might be caused by the limitation of individual studies. To shed light on the associations between the RNASEL gene polymorphism and prostate cancer risk, we performed this meta-analysis of 16 case–control studies retrieved from 13 published papers.7–19
Methods
Search strategy
We searched the PubMed, Embase, and Web of Science databases for papers published in the past 20 years from 1997 through to 2017, using the keywords “RNASEL”, “cancer or carcinoma or tumor”, and “polymorphism or variant”. A search conducted with these terms yielded a total of 81 publications, of which 14 were finally selected. We also screened the referenced literature of the selected publications manually.
Inclusion and exclusion criteria
Studies were required to fulfill the following inclusion criteria: they should 1) assess the correlation between prostate cancer risk and the RNASEL −1385G/A polymorphism, 2) be case–control studies, and 3) consider sufficient numbers of genotypes for cases and controls. Studies were excluded if they 1) included no control population, 2) did not report genotype frequency data, and 3) were publications using the same dataset.
Data extraction
Two authors independently extracted all data from the selected publications. These data included the first author’s last name, year of publication, country of origin, ethnicity of the study population, total number of samples in case and control groups, numbers of each genotype in both case and control groups, P-value of Hardy–Weinberg equilibrium (HWE) of the control group, and genotyping method. Ethnicity was categorized as individuals of European descent and African or Asian, and control subgroups were population-based and hospital-based (HB).
Statistical analysis
Based on genotype frequencies for cases and controls, crude odds ratio (OR) with 95% confidence interval (CI) was used to measure the strength of associations between the rs486907 polymorphism and prostate cancer risk. The statistical significance of the OR was determined with the Z test. The heterogeneity assumption among studies was evaluated using a χ2-based Q test. A P-value of >0.10 for the Q test indicated a lack of heterogeneity among studies. If significant heterogeneity was detected, the DerSimonian–Laird random-effects model or the Mantel–Haenszel fixed-effects model was chosen for analysis.20,21 We investigated the relationship between the rs486907 genetic variant of the RNASEL gene and cancer risk by testing for the allelic contrast (G vs A), homozygote comparison (GG vs AA), heterozygote comparison (GG vs GA), and dominant genetic model (GG + GA vs AA). A sensitivity analysis was performed by omitting studies, one after the other, to assess the stability of results. The departure of frequencies of the RNASEL polymorphism from expectations under HWE was assessed by the Pearson’s χ2 test, and the values of P<0.05 were considered significant.22 Publication bias was investigated using Egger’s linear regression method and Begg’s funnel plots, where P<0.05 was considered significant.23 All statistical tests for meta-analyses were performed using Stata software version 10.0 (StataCorp LP, College Station, TX, USA).
Results
Study characteristics
Using various combinations of key terms, a total of 81 publication titles were identified from the PubMed, Embase, and Web of Science databases. As shown in Figure 1, 40 publications were excluded from the analysis after screening the abstracts. The full texts of the remaining papers were then evaluated, and 27 additional publications were excluded for the following reasons: duplications (10), meta-analyses or systematic reviews (1), publications dealing with other polymorphisms (16), and one publication having no case–control group. Finally, 13 publications including 16 case–control studies were chosen for our meta-analysis. The distribution of genotypes in all the control groups was in agreement with HWE. The genotyping methods used in the different studies that were a part of our meta-analysis included restrictive fragment length polymorphism polymerase chain reaction, TaqMan, Sequenom platform, and mass spectrometry-based genotyping analysis. Characteristics of the case–control studies included in this meta-analysis are shown in Table 1.
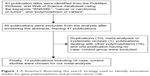  | Figure 1 A flowchart illustrating the search strategy used to identify association studies for gene polymorphisms and prostate cancer risk. |
Quantitative synthesis
Results of the overall meta-analysis are shown in Table 2. There was no positive association between this polymorphism and prostate cancer susceptibility (Figure 2). When stratified by ethnicity, we observed that the polymorphism was associated with a decreased prostate cancer risk in African-descent patients (GG/GA vs AA: OR =0.366, 95% CI =0.174–0.770, Pheterogeneity =0.095; GG vs AA: OR =0.370, 95% CI =0.176–0.783, Pheterogeneity =0.098; Figure 3). Analysis of the subgroups based on the source of control indicated that the polymorphism was associated with a decreased prostate cancer risk in HB controls (GG vs AA: OR =0.697, 95% CI =0.488–0.996, Pheterogeneity =0.000; GG + GA vs AA: OR =0.701, 95% CI =0.502–0.978, Pheterogeneity =0.001; Figure 4).
  | Figure 2 Forest plot of the rs486907 polymorphism and prostate cancer risk: GG vs AA for total. |
  | Figure 3 Forest plot of the rs486907 polymorphism and prostate cancer risk: GG + GA vs AA for Africans. |
Bias diagnosis and sensitivity analysis
Egger’s and Begg’s tests were performed to assess publication bias. No proof of publication bias was found, except overall GG vs GA and GG/GA vs AA. A sensitivity analysis was performed to assess the influence of each study on the pooled OR by sequential removal of individual studies. Neither Begg’s funnel plot nor Egger’s test detected any obvious evidence of publication bias in the subgroup analyses for all genetic models. Figure 5 shows the funnel plot for GG vs AA of prostate cancer in Africans.
  | Figure 5 Begg’s funnel plot for publication bias test of prostate cancer (GG vs AA). |
Discussion
Under normal conditions, RNASEL has tumor-suppressive and antiproliferative functions, although the common rs486907 variant of RNASEL gene has been associated with risk of a number of cancers.21–24 Up to now, many molecular epidemiological studies were performed to assess the associations of this polymorphism with prostate cancer risk. In a large American series, Casey et al observed statistically significant ORs for prostate cancer patients carrying the RNASEL variant G-allele (GG vs AA: OR =2.12, 95% CI =1.19–3.78; GA vs AA: OR =1.46, 95% CI =0.65–1.19).6 In 2002, Rökman et al analyzed 233 unselected patients with prostate cancer and 176 unaffected sibling controls from Finland, and found that the Gln/Gln variant was more frequent in 66 patients with hereditary prostate cancer than in 176 controls, but was not more frequent in unselected 167 patients with prostate cancer compared with controls.26 By contrast, Wang et al found that homozygosity for the Arg variant was less frequent in 493 controls compared with 433 familial prostate cancer patients from the USA.17 Following these studies, two more papers on the association involving larger sample sizes were published,24,25 which indicated that the rs486907 polymorphism in the RNASEL gene is not associated with an increased risk of developing prostate cancer. In order to verify this association, our meta-analysis combined data from 7,602 patients with prostate cancer and 7,108 controls, and revealed that the rs486907 polymorphism was not associated with prostate cancer risk. When stratified by ethnicity, we found that the rs486907 variant was associated with a decreased cancer risk in Africans (GG vs AA: OR =0.370, 95% CI =0.176–0.783; GG/GA vs AA: OR =0.366, 95% CI =0.174–0.770). Therefore, the variant G-allele might be a protective factor against prostate cancer in African-descent individuals.
Although we expended considerable efforts in identifying as many associations between the RNASEL gene polymorphism and cancer risk as possible, our study still suffers from some inherent limitations. First, the different genotyping strategies may contribute to bias in the analysis. Second, factors associated with gene–gene and gene–environment interactions were poorly represented. It is possible that specific environmental and lifestyle factors such as age, sun exposure, smoking, drinking, and family history could alter the associations between the gene polymorphism and prostate cancer risk. Therefore, it is necessary to determine the roles of specific environmental factors and lifestyles. Third, we were unable to analyze how the different developmental stages of prostate cancer are associated with the RNASEL polymorphism.
In conclusion, our present meta-analysis suggests that the rs486907 polymorphism in the RNASEL gene may not be associated with prostate cancer risk. However, the variant G allele might be a protective factor against prostate cancer in African-descent individuals. Further studies based on larger sample sizes and considering additional gene–environment interactions should be conducted in the future.
Disclosure
The authors report no conflicts of interest in this work.
References
Rusch L, Zhou A, Silverman RH. Caspase-dependent apoptosis by 29,59-oligoadenylate activation of RNase L is enhanced by IFN-beta. J Interferon Cytokine Res. 2000;20(12):1091–1100. | ||
Silverman RH. Implications for RNase L in prostate cancer biology. Biochemistry. 2003;42(7):1805–1812. | ||
Ezelle HJ, Hassel BA. Pathologic effects of RNase-L dysregulation in immunity and proliferative control. Front Biosci (Schol Ed). 2012;4:767–786. | ||
Brennan-Laun SE, Ezelle HJ, Li XL, Hassel BA. RNase-L control of cellular mRNAs: roles in biologic functions and mechanisms of substrate targeting. J Interferon Cytokine Res. 2014;34(4):275–288. | ||
Banerjee S, Li G, Li Y, et al. RNase L is a negative regulator of cell migration. Oncotarget. 2015;6(42):44360–44372. | ||
Casey G, Neville PJ, Plummer SJ, et al. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet. 2002;32(4):581–583. | ||
Alvarez-Cubero MJ, Pascual-Geler M, Martinez-Gonzalez LJ, et al. Association between RNASEL, MSR1, and ELAC2 single nucleotide polymorphisms and gene expression in prostate cancer risk. Urol Oncol. 2016;34(10):431.e1–431.e8. | ||
Cybulski C, Wokołorczyk D, Jakubowska A, et al. DNA variation in MSR1, RNASEL and E-Cadherin genes and prostate cancer in Poland. Urol Int. 2007;79(1):44–49. | ||
Daugherty SE, Hayes RB, Yeager M, et al. RNASEL Arg462Gln polymorphism and prostate cancer in PLCO. Prostate. 2008;67(8):849–854. | ||
Nam RK, Zhang WW, Jewett MA, et al. The use of genetic markers to determine risk for prostate cancer at prostate biopsy. Clin Cancer Res. 2010;11(23):8391–8397. | ||
Beuten J, Gelfond JA, Franke JL, et al. Single and multivariate associations of MSR1, ELAC2, and RNASEL with prostate cancer in an ethnic diverse cohort of men. Cancer Epidemiol Biomarkers Prev. 2010;19(2):588–599. | ||
Robbins CM, Hernandez W, Ahaghotu C, et al. Association of HPC2/ELAC2 and RNASEL non-synonymous variants with prostate cancer risk in African American familial and sporadic cases. Prostate. 2008;68(16):1790–1797. | ||
Wiklund F, Jonsson BA, Brookes AJ, et al. Genetic analysis of the RNASEL gene in hereditary, familial, and sporadic prostate cancer. Clin Cancer Res. 2004;10(21):7150–7156. | ||
Nakazato H, Suzuki K, Matsui H, Ohtake N, Nakata S, Yamanaka H. Role of genetic polymorphisms of the RNASEL gene on familial prostate cancer risk in a Japanese population. Br J Cancer. 2003;89(4):691–696. | ||
Rökman A, Ikonen T, Seppälä EH, et al. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am J Hum Genet. 2002;70(5):1299–1304. | ||
Maier C, Haeusler J, Herkommer K, et al. Mutation screening and association study of RNASEL as a prostate cancer susceptibility gene. Br J Cancer. 2005;92(6):1159–1164. | ||
Wang L, McDonnell SK, Elkins DA, et al. Analysis of the RNASEL gene in familial and sporadic prostate cancer. Am J Hum Genet. 2002;71(1):116–123. | ||
Shea PR, Ishwad CS, Bunker CH, Patrick AL, Kuller LH, Ferrell RE. RASEL and RNASEL-inhibitor variation and prostate cancer risk in Afro-Caribbeans. Prostate. 2008;68(4):354–359. | ||
Shook SJ, Beuten J, Torkko KC, et al. Association of RNASEL variants with prostate cancer risk in Hispanic Caucasians and African Americans. Clin Cancer Res. 2007;13(19):5959–5964. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Napolioni V. The relevance of checking population allele frequencies and Hardy-Weinberg Equilibrium in genetic association studies: the case of SLC6A4 5-HTTLPR polymorphism in a Chinese Han Irritable Bowel Syndrome association study. Immunol Lett. 2014;162(1 Pt A):276–278. | ||
Hayashino Y, Noguchi Y, Fukui T. Systematic evaluation and comparison of statistical tests for publication bias. J Epidemiol. 2005;15(6):235–243. | ||
Andersen JB, Li XL, Judge CS, et al. Role of 2–5A-dependent RNase-L in senescence and longevity. Oncogene. 2007;26(21):3081–3088. | ||
Castelli JC, Hassel BA, Wood KA, et al. A study of the interferon antiviral mechanism: apoptosis activation by the 2–5A system. J Exp Med. 1997;186(6):967–972. | ||
Rökman A, Ikonen T, Seppälä EH, et al. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am J Hum Genet. 2002;70(5):1299–1304. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



