Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 13
The Relationship Between Single Nucleotide Polymorphisms of SMAD3/SMAD6 and Risk of Esophageal Squamous Cell Carcinoma in Chinese Population
Authors Yu J , Dong Y, Tang W, Pan H, Lv L, Long T, Zhou Q, Qi J, Liu J, Ding G, Yin J, Tan L
Received 16 February 2020
Accepted for publication 20 July 2020
Published 24 August 2020 Volume 2020:13 Pages 355—363
DOI https://doi.org/10.2147/PGPM.S250076
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Jinjie Yu,1,* Yunpeng Dong,1,* Weifeng Tang,2 Huiwen Pan,2 Lu Lv,2 Tao Long,2 Qiang Zhou,2 Junqing Qi,2 Jianchao Liu,2 Guowen Ding,2 Jun Yin,1 Lijie Tan1
1Department of Thoracic Surgery, Zhongshan Hospital of Fudan University, Shanghai, People’s Republic of China; 2Department of Cardiothoracic Surgery, Affiliated People’s Hospital of Jiangsu University, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jun Yin; Lijie Tan
Department of Thoracic Surgery, Zhongshan Hospital of Fudan University, No. 180, Fenglin Road, Xuhui District, Shanghai, People’s Republic of China
Tel +86 13917483128
; +86 13681972151
Email [email protected]; [email protected]
Background: The TGF-β signal pathways play a key role in the development and promotion of squamous cell carcinoma (SCC). The pathway is mediated by the SMAD family proteins that include SMAD3 and SMAD6. Our study aimed to evaluate the relationship between single nucleotide polymorphism (SNP) of SMAD3/SMAD6 and susceptibility to esophageal squamous cell carcinoma (ESCC) in the Chinese population.
Patients and Methods: This was a hospital-based case–control study compromised of 1043 ESCC patients and 1315 non-cancer patients. Seven SMAD3/SMAD6 (rs8028147, rs3743343, rs3743342, rs8025774, rs8031440, rs803167, and rs34643453) SNPs were selected and used to evaluate their correlation with ESCC susceptibility. Genetic model tests, stratified analyses, linkage disequilibrium analyses, and haplotype analyses were performed in our study.
Results: Participants with SMAD3 rs3743342 C>T, rs8025774 C>T, rs8031440 G>A or rs8031627 G>A had a significantly higher risk of ESCC. This was more evident in males, older patients (> 63 years), smokers, and non-alcohol drinking participants. Linkage disequilibrium analyses further revealed that there were strong correlations between SMAD3 rs3743342 C>T, rs8025774 C>T, rs8031440 G>A, and rs8031627 G>A. In the same line, haplotype analyses revealed that SMAD3 ACCCGGSMAD6A and SMAD3AGCCGGSMAD6A were associated with less susceptibility to ESCC while SMAD3ATTTAASMAD6A was associated with a higher risk of ESCC.
Conclusion: SNPs of SMAD3 were related to higher susceptibility to ESCC. As such, they may contribute to the development of viable strategies for early diagnosis and treatment of ESCC. However, more detailed association mechanisms between SMAD3/SMAD6 SNPs and ESCC need further experiments to prove.
Keywords: SMAD, single nucleotide polymorphism, esophageal squamous cell carcinoma
Introduction
Small mothers against decapentaplegic (SMAD) proteins are a family of transducers and transcriptional modulators that functions by transforming the growth factor beta (TGF-β) signaling pathway. Alterations of the TGF-β signal pathway lead to uncontrolled cell proliferation, thereby contributing to oncogenesis.1 Ligands of the TGF-β family bind to complexes formed by its type I and type II receptors to trigger the pathway. The activated type I receptors then phosphorylate receptor-activated SMADs (R-SMADs, including SMAD2, −3, −5, −8) which then interact with SMAD4 (also known as the co-SMAD) to form a heterotrimeric complex that consists of two R-SMADs and one co-SMAD.1,2 A subgroup of inhibitory SMADs (I-SMADs, including SMAD6, −7) attenuates the signaling by competing with the R-SMADs for TGF-β receptors or directly interacting with R-SMADs.1,3,4
Drugs that mediate the TGF-β signaling pathway have shown increasing potency of changing the current treatment of various cancers.5 Given that the SMADs family play a key role in activation or inhibition of the TGF-β signaling pathway, further studies on SMADs are becoming essential and promising.6 SMAD3 has been proven to have diverse correlations with different types of malignant tumors. For example, SMAD3 impairment is associated with dysregulated cell proliferation and apoptosis in ovarian granulosa cell tumors.7 In human colorectal cancer, linker phosphorylation of SMAD3 is associated with tumor metastasis.8 Inhibition of SMAD3 diminishes the invasion and metastatic ability of breast cancer.9,10 In addition, the expression of SMAD3 suppressed tumor development in gastric cancer.11 Similarly, SMAD6 has been demonstrated to be related to poor prognosis in non-small cell lung cancer.12,13 However, there is only little evidence that shows SMAD3 or SMAD6 contribute to the progression of ESCC, especially the relationship between SMAD gene and ESCC. Cognizant to this, our study aimed to explore whether SNPs in SMAD3 or SMAD6 had an effect on the risk of ESCC.
Patients and Methods
Ethical Statement
This was a case–control study approved by the Ethics Review Board of Jiangsu University (Zhenjiang, China). All study participants gave written informed consent prior to the study. The study also complied with the World Medical Association Declaration of Helsinki regarding ethical aspects of research involving human/animal subjects.
Study Population
A total of 2358 participants were enrolled in the study, of which 1043 were ESCC cases and 1315 non-cancer individuals acted as the controls. Frequency-matching of both groups was done based on gender and age. All participants were consecutively recruited from the Affiliated People’s Hospital of Jiangsu University and Affiliated Hospital of Jiangsu University (Zhenjiang, China) between October 2008 and January 2017. All cases of ESCC were diagnosed histologically. Patients with a history of malignant tumor, metastasized cancer, and/or those who had undergone chemotherapy/radiotherapy were excluded from the study.
Individual interviews were conducted using a questionnaire well formulated to collect data of all the necessary demographic information and related risk factors. Then, participants gave 2 mL venous blood samples for subsequent tests. Participants who smoked at least one cigarette per day for the last one year or more were categorized as the “tobacco consumption” subgroup while those who drank more than three alcoholic drinks per week for the past six months or more were categorized as the “alcohol consumption” subgroup.
DNA Extraction and SNP Genotyping
Genomic DNA was isolated from the collected venous blood samples using the QIAamp DNA Blood Mini Kit (Qiagen, Berlin, Germany) following the manufacturer’s protocol.14 The genotypes of 7 SNPs were then analyzed using the ligation detection reaction (LDR) method with technical support from Genesky Biotechnology Inc. (Shanghai, China). Quality control was done by repeating the analyses using 10% of randomly selected samples. Pilot linkage disequilibrium analyses were performed in the Chinese Han population to choose the SNP loci with moderate correlation, and tag SNPs were selected for further analyses.
Statistical Analysis
All statistical analyses were conducted using the SPSS 25.0 statistical software package (SPCC Inc., Chicago, IL). Hardy–Weinberg equilibrium (HWE) for the genotypes was tested by goodness-of-fit χ2 in the control group. Variations in demographic characteristics and genotypes of the SMAD3 rs8028147, rs3743343, rs3743342, rs8025774, rs8031440, rs8031627, and SMAD6 rs34643453 between both groups were evaluated using the χ2 test to determine their statistical differences. The associations between these 7 SNPs and risk of ESCC were assessed by odds ratio (OR) and 95% confidence interval (CI) using logistics regression analyses for crude ORs and adjusted ORs according to age, sex, and tobacco and alcohol consumption status. Two-sided P value <0.05 was considered as statistically significant.
Results
Characteristics of the Study Population
The demographic information and risk factors of the participants are shown in Table 1. Age and gender were well matched in cases and control groups. However, the smoking and alcohol drinking statuses were significantly different between the groups (P<0.01).
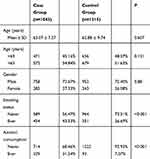 |
Table 1 Distribution of Selected Demographic Variables and Risk Factors in ESCC Case and Control Groups |
Table 2 provides primary information of the 7 genotyped SNPs. The success rates of SNP genotyping were variable and the rates ranged between 95% and 100%. The minor allele frequency (MAF) of SNPs in the control group corresponded with that of the Chinese Han population provided by HapMap. Deviation tests for the HWE revealed that the control population was in the Hardy–Weinberg proportions for all the 7 SNPs with a significance level of 0.05.
 |
Table 2 Primary Information of the Selected SNPs |
The Risk of ESCC Associated with SNPs
The association between the risk of ESCC and each SNP is presented in Table 3. It presents the results obtained from the analysis of the association between the risk of ESCC and each SNP. The co-dominant model test, dominant model test, recessive model test, and an allelic test revealed that among the selected SNPs, SMAD3 rs3743342 C>T, rs8025774 C>T, rs8031440 G>A, and rs8031627 G>A were closely associated with a higher risk of ESCC in the case groups (P<0.05).
 |
Table 3 Main Effects of Rs8028147 G>A, Rs3743343 T>C, Rs3743342 C>T, Rs8025774 C>T, Rs8031440 G>A, Rs8031627 G>A, Rs3463453 G>A on Risk of ESCC |
Stratification Analyses of SMAD3 rs3743342 C>T, rs8025774 C>T, rs8031440 G>A, rs8031627 G>A and the Risk of ESCC
Stratification analyses were conducted to further access SMAD3 rs3743342 C>T, rs8025774 C>T, rs8031440 G>A, and rs8031627 G>A on the risk of ESCC in the different subgroups based on gender, age, and smoking and alcohol drinking status.
The results of stratification analyses share a remarkable similarity (Tables 4–7). In males, participants older than 63 years, smokers, and non-alcohol drinking participants, almost all the mutant homozygotes of these four SNPs (except rs8031440 G>A in smokers) had a significantly higher likelihood of having ESCC (P<0.05).
 |
Table 4 Stratified Analyses Between Rs3743342 Polymorphism and ESCC Risk by Gender, Age, Smoking Status, and Alcohol Consumption |
 |
Table 5 Stratified Analyses Between Rs8025774 Polymorphism and ESCC Risk by Gender, Age, Smoking Status, and Alcohol Consumption |
 |
Table 6 Stratified Analyses Between Rs8031440 Polymorphism and ESCC Risk by Gender, Age, Smoking Status, and Alcohol Consumption |
 |
Table 7 Stratified Analyses Between Rs8031627 Polymorphism and ESCC Risk by Gender, Age, Smoking Status, and Alcohol Consumption |
Linkage Disequilibrium Analyses and Association Test
Linkage disequilibrium analyses of both the control and case groups are set out in supplementary Tables 1 and 2. There were significant correlations between SMAD3 rs3743342 C>T, rs8025774 C>T, rs8031440 G>A and rs8031627 G>A and risk of ESCC.
Haplotype Analysis of Polymorphisms and Susceptibility to ESCC
As summarized in Table 8, haplotype analysis of 7 SNPs showed that SMAD3Ars8028147Crs3743343Crs3743342Crs8025774Grs8031440Grs8031627SMAD6Grs3463453 (OR=0.809, 95% CI=0.674–0.972, P=0.023) and SMAD3 Ars8028147Grs3743343Crs3743342Crs8025774Grs8031440Grs8031627SMAD6Ars3463453 (OR=0.569, 95% CI=0.395–0.820, P=0.002) were associated with less susceptibility to ESCC, while SMAD3Ars8028147Trs3743343Trs3743342Trs8025774Ar8031440Ars8031627SMAD6Ars3463453 was associated with higher risk of ESCC (OR = 1.318, 95% CI=1.056–1.645, P= 0.014).
 |
Table 8 Haplotype Frequencies in the Case and Control Group, and Risk of ESCC |
Discussion
Herein, the association between SMAD3/SMAD6 SNPs and the risk of ESCC among the Chinese population was assessed. Preliminary analysis revealed that the distributions of the 7 SNPs were consistent with that of HapMap data. As such, the results of our study could be generalized and used for the entire Chinese Han population.
Several studies postulate that SMAD3 plays a protective role in ESCC. Our study showed that the association between SMAD3 rs3743342 C>T, rs8025774 C>T, rs8031440 G>A, rs8031627 G>A, and ESCC was consistent in different genetic models. Thus, selected SMAD3 SNPs may affect the susceptibility of the participants to ESCC in a positive correlated manner. However, mutant heterozygote of the mentioned 4 SNPs seemed to have no significant association with risk of ESCC compared with the wild genotype in the co-dominant model, which may be due to the inheritance mode and mechanism of SNPs on tumor development and progression.
Stratification analyses of the four SNPs further revealed that their effects varied in different subgroups. The SNPs significantly increased the risk of ESCC in males and participants aged more than 63 years. Smokers with SMAD3 rs3743342 C>T, rs8025774 C>T, or rs8031627 G>A were more susceptible to ESCC while those with the rs8031440 wild-type homozygotes had a lower risk of ESCC than those with other genotypes. These results were consistent with previous results that showed ESCC is more prevalent in Chinese males than females, and in elder people than younger people, and smoking increases the risk of ESCC by about 3–7-fold.15,16 Cognizant to this, these results suggested that there exists an interaction between the environmental and genetic risk factors in tumorigenesis of ESCC. Non-alcohol drinking participants with SMAD3 rs3743342 C>T, rs8025774 C>T, rs8031440, or rs8031627 G>A were more susceptible to ESCC. However, there was no such correlation in the alcohol-drinking subgroup. The result seemed contradictory to the evidence that alcohol drinking is a significant contributory factor to the development of ESCC.17 Thus, the mechanism underlying this discrepancy should be further investigated.
There was a significant correlation between rs3743342, rs8025774, rs8031440, and rs8031627 that further confirmed their similarities. The four SNPs were all located in the three-prime untranslated region (3ʹ-utr) of SMAD3. Many recent studies have reported that the 3ʹ-utr of SMAD3 has an important impact on the development of various malignant tumors. For example, inhibition of micro-RNAs that target at the 3ʹ-utr of SMAD3 leads to the upregulation of SMAD3, thereby constraining the epithelial–mesenchymal transition and invasion of non-small cell lung cancer.18 In the same line, silencing the micro-RNAs that target the 3ʹ-utr of SMAD3 decreases the expression of SMAD3, thereby inhibiting the proliferation of glioblastoma cells.19 Based on these reports, it appears that these SNPs influence the risk of ESCC through post-transcriptional regulation. Raine et al reported that rs8031440 and rs3743342 were also correlated with primary osteoarthritis, aneurysms, and osteoarthritis syndrome.20 It can be reasonably assumed that these SNPs affect the susceptibility to ESCC as well as that to other diseases.
The major limitation of our study was the lack of technical support to establish a single nucleotide mutation cell or animal model. As such, the biological function of these SNPs requires further research. In addition, our study was conducted in a single center, although the sample size was impressive.
Conclusion
SMAD3 rs3743342 C>T, rs8025774 C>T and rs8031627 G>A increase the susceptibility of individuals to ESCC, particularly in males, people aged over 63 years, smokers, and non-alcohol drinking people. The distribution of the 7 SNPs was consistent with that of HapMap data based on their primary data. As such, the results of this study can be generalized and used as a useful resource for ESCC screening of the entire Chinse Han population.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
All authors have no conflicts of interest to declare.
References
1. Budi EH, Duan D, Derynck R. Transforming growth factor-β receptors and SMADs: regulatory complexity and functional versatility. Trends Cell Biol. 2017;27:658–672. doi:10.1016/j.tcb.2017.04.005
2. Macias MJ, Martin-Malpartida P, Massagué J. Structural determinants of SMAD function in TGF-β signaling. Trends Biochem Sci. 2015;40:296–308. doi:10.1016/j.tibs.2015.03.012
3. Miyazawa K, Miyazono K. Regulation of TGF-β family signaling by inhibitory SMADs. CSH Perspect Biol. 2017;9:a22095.
4. Chen W, Ten Dijke P. Immunoregulation by members of the TGFβ superfamily. Nat Rev Immunol. 2016;16:723–740. doi:10.1038/nri.2016.112
5. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019;18:197–218. doi:10.1038/s41573-018-0007-y
6. Zhu H, Gu X, Xia L, et al. A novel TGFβ trap blocks chemotherapeutics-induced TGFβ1 signaling and enhances their anticancer activity in gynecologic cancers. Clin Cancer Res. 2018;24:2780–2793. doi:10.1158/1078-0432.CCR-17-3112
7. Fang X, Gao Y, Li Q. SMAD3 activation: a converging point of dysregulated TGF-beta superfamily signaling and genetic aberrations in granulosa cell tumor development? Biol Reprod. 2016;95:105. doi:10.1095/biolreprod.116.143412
8. Matsuzaki K, Kitano C, Murata M, et al. SMAD2 and SMAD3 phosphorylated at both linker and COOH-terminal regions transmit malignant TGF-β signal in later stages of human colorectal cancer. Cancer Res. 2009;69:5321–5330. doi:10.1158/0008-5472.CAN-08-4203
9. Bae E, Sato M, Kim RJ, et al. Definition of SMAD3 phosphorylation events that affect malignant and metastatic behaviors in breast cancer cells. Cancer Res. 2014;74:6139–6149. doi:10.1158/0008-5472.CAN-14-0803
10. Zhao Y, Ma J, Fan Y, et al. TGF-beta transactivates EGFR and facilitates breast cancer migration and invasion through canonical SMAD3 and ERK/Sp1 signaling pathways. Mol Oncol. 2018;12:305–321. doi:10.1002/1878-0261.12162
11. Han S, Kim H, Seong DH, et al. Loss of the SMAD3 expression increases susceptibility to tumorigenicity in human gastric cancer. Oncogene. 2004;23:1333–1341. doi:10.1038/sj.onc.1207259
12. Cho SY, Ha SY, Huang S, et al. The prognostic significance of SMAD3, SMAD4, SMAD3 phosphoisoform expression in esophageal squamous cell carcinoma. Med Oncol. 2014;31. doi:10.1007/s12032-014-0236-9
13. Jeon HS, Dracheva T, Yang SH, et al. SMAD6 contributes to patient survival in non-small cell lung cancer and its knockdown reestablishes TGF- homeostasis in lung cancer cells. Cancer Res. 2008;68:9686–9692. doi:10.1158/0008-5472.CAN-08-1083
14. Yin J, Wang L, Tang W, et al. RANK rs1805034 T>C polymorphism is associated with susceptibility of esophageal cancer in a Chinese population. PLoS One. 2014;9:e101705. doi:10.1371/journal.pone.0101705
15. Zheng R, Zeng H, Zhang S, Chen T, Chen W. National estimates of cancer prevalence in China, 2011. Cancer Lett. 2016;370:33–38. doi:10.1016/j.canlet.2015.10.003
16. Kamangar F, Chow W, Abnet C, Dawsey S. Environmental causes of esophageal cancer. Gastroenterol Clin North Am. 2009;38:27–57. doi:10.1016/j.gtc.2009.01.004
17. Salaspuro MP. Alcohol consumption and cancer of the gastrointestinal tract. Best Pract Res Clin Gastroenterol. 2003;17:679–694. doi:10.1016/S1521-6918(03)00035-0
18. Hu H, Xu Z, Li C, et al. MiR-145 and miR-203 represses TGF-β-induced epithelial-mesenchymal transition and invasion by inhibiting SMAD3 in non-small cell lung cancer cells. Lung Cancer. 2016;97:87–94. doi:10.1016/j.lungcan.2016.04.017
19. Wu ZB, Cai L, Lin SJ, Lu JL, Yao Y, Zhou LF. The miR-92b functions as a potential oncogene by targeting on SMAD3 in glioblastomas. Brain Res. 2013;1529:16–25. doi:10.1016/j.brainres.2013.07.031
20. Raine EVA, Reynard LN, van de Laar IMBH, Bertoli-Avella AM, Loughlin J. Identification and analysis of a SMAD3 cis-acting eQTL operating in primary osteoarthritis and in the aneurysms and osteoarthritis syndrome. Osteoarthritis Cartil. 2014;22:698–705. doi:10.1016/j.joca.2014.02.931
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
