Back to Journals » OncoTargets and Therapy » Volume 9
The relationship between apolipoprotein E gene ε2/ε3/ε4 polymorphism and breast cancer risk: a systematic review and meta-analysis
Authors Liu Y, Zhang H, Pan H, Bao Y, Xue J, Wang T, Dong X, Li X, Bao H
Received 11 August 2015
Accepted for publication 21 November 2015
Published 7 March 2016 Volume 2016:9 Pages 1241—1249
DOI https://doi.org/10.2147/OTT.S94228
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Faris Farassati
Yun-Long Liu,1 Hao-Min Zhang,1 Hong-Ming Pan,2 Yu-Hang Bao,2 Jing Xue,2 Tian-Chang Wang,1 Xiao-Cheng Dong,1 Xiao-Ling Li,3 Hong-Guang Bao1
1Department of Chest Surgery, The Second Affiliated Hospital of Qiqihar Medical University, 2Basic Medical Science College, Qiqihar Medical University, Qiqihar, Heilongjiang, People’s Republic of China; 3Department of Anatomy, Basic Medical Science College, Qiqihar Medical University, Qiqihar, Heilongjiang, People’s Republic of China
Objective: We conducted a systematic review and meta-analysis aiming to assess the relationship between apolipoprotein E (APOE) gene ε2/ε3/ε4 polymorphism and breast cancer risk.
Methods: Yun-Long Liu and Hao-Min Zhang independently completed literature retrieval and data collection, and statistical analyses were performed by Stata. Individual odds ratio (OR) and 95% confidence interval (CI) were pooled in a random-effects model using the DerSimonian–Laird method. Heterogeneity was evaluated by I2 statistic at a significance level of 50%. Publication bias was assessed by Egger’s test.
Results: Eleven articles including 2,074 breast cancer patients and 2,372 controls were summarized. Using the most common allele ε3 as a reference, the ε2 (OR =0.87, 95% CI =0.72–1.05, P=0.154, I2=0.0%) and ε4 (OR =1.07, 95% CI =0.80–1.42, P=0.654, I2=71.8%) alleles were not found to be significantly associated with breast cancer risk in the overall analyses. Subgroup analyses revealed that the comparison of allele ε4 with ε3 was significant in Asians (OR =1.58, 95% CI =1.17–6.32, P=0.003, I2=12.1%) and in studies that used the restriction fragment length polymorphism (RFLP) genotyping method (OR =1.27; 95% CI =1.01–1.61, P=0.045, I2=34.3%), and was marginally significant in hospital-based studies (OR =1.33; 95% CI =0.98–1.79, P=0.065, I2=30.2%), without heterogeneity. Moreover, the presence of the ε2 allele was significantly associated with breast cancer in small studies (total sample size <500) (OR =0.73, 95% CI =0.54–1.00, P=0.052, I2=0.0%) without heterogeneity. The Egger’s test indicated low probabilities of publication bias.
Conclusion: We observed a significant association between APOE gene ε4 allele and breast cancer risk in Asian populations. Moreover, the findings of our subgroup analyses suggest that source of controls, genotyping platform, and sample size might be the potential causes of heterogeneity.
Keywords: apolipoprotein E, breast cancer, polymorphism, meta-analysis
Introduction
Breast cancer is a heterozygous disease with a strong heritable component, and it is estimated that ~15% of breast cancer cases have at least one affected first-degree relative.1 Despite the tremendous advancement gained in medical sciences, to unravel the genetic architecture of breast cancer still remains a challenge. So far, a number of susceptibility genes have been suggested to underlie breast carcinogenesis.2,3 One of the promising candidate genes is apolipoprotein E (APOE) and its expression is proposed as a tumor-associated biomarker.4 APOE is a class of apolipoprotein and its primary function is to mediate cholesterol metabolism and catabolize triglyceride-rich lipoprotein constituents. Beyond this well-known function, evidence is amassing to reveal a different role for APOE in carcinogenesis and tumor development.5 The gene encoding APOE is mainly inherited as one of the three alleles: ε2 (Arg148→Cys), ε3 (Cys112), and ε4 (Cys112→Arg), which generate three homozygous genotypes (ε2/2, ε3/3, and ε4/4) and three heterozygous genotypes (ε2/3, ε2/4, and ε3/4), each with a diverse receptor-binding capacity.6 In addition, there are several other minor alleles such as ε5 and ε7 in APOE gene; due to the methodological drawbacks of restriction fragment length polymorphism (RFLP), these minor alleles could be easily missed.7,8 In medical research literature, the ε2, ε3, and ε4 alleles in APOE gene are given the most attention, and their susceptibility toward breast cancer risk has been widely evaluated by genetic association studies;9–11 however, the results of only a few studies are reproducible. A previous meta-analysis of eight articles by Saadat12 found that APOE gene ε4 allele or ε4 positive genotypes was exclusively associated with breast cancer in Asians. Further meta-analysis by Anand et al13 failed to validate this significant finding for ε4 positive genotypes in overall susceptibility to breast cancer. Besides unavoidable genetic heterogeneity across races and ethnicities, a possible explanation for such divergence is inadequate statistical rigor to quantify the effect size reliably. To obtain a more precise estimate, we conducted an updated systematic review and meta-analysis of previous studies by incorporating additional articles and seeking other potential causes of heterogeneity in both subgroup and meta-regression analyses.
Methods
This systematic review and meta-analysis was conducted based on the guidelines listed in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.
Literature retrieval
A comprehensive literature search was conducted in the Medline, Embase, and Web of Science electric databases. Key subjects used for retrieval included apolipoprotein E or APOE or APO E and breast cancer or breast carcinoma. The last search was on July 5, 2015. Each retrieved article was reviewed from the title or abstract to definitively exclude the one with irrelevant content. In case of uncertainty, the full text was downloaded for further review. To avoid missing hits, the bibliographies of original articles and reviews were manually searched.
Eligibility appraisal
The eligibility of each retrieved article was appraised according to the following criteria: 1) the clinical endpoint must be breast cancer with validated clinical diagnoses; 2) it should be designed as a case–control association study; 3) the genotypes or alleles of APOE gene ε2/ε3/ε4 polymorphism or its associated risk estimates – odds ratio (OR) or beta and its 95% confidence interval (95% CI) or standard error – should be provided; and 4) conference abstracts or proceedings (due to insufficient necessary information) and non-English articles are not considered. In case of more than one independent study in a single article, each study was summarized and analyzed separately.
Information extraction
Relevant information from each eligible article was extracted separately by two authors (Yun-Long Liu and Hao-Min Zhang) according to a predefined table designed by all responsible authors, including authors, the year of publication, race/ethnicity, study design, source of controls, genotyping method, sample size, age, body mass index (BMI), and the genotype or allele counts of APOE gene ε2/ε3/ε4 polymorphism or the associated ORs and 95% CIs. In case of unresolved-by-discussion disagreements, a third senior author (Hong-Guang Bao) was involved until a consensus was reached.
Statistical analysis
The risk estimate for APOE gene ε2/ε3/ε4 polymorphism in susceptibility to breast cancer was expressed as OR and its 95% CI, which were pooled in a random-effects model using the method proposed by DerSimonian and Laird.14 The heterogeneity among the results of different studies was weighed by the I2 statistic, which is defined as the percentage of observed between-study variability that stems from heterogeneity rather than chance. As recommended by Higgins et al, an I2 statistic of 50% or greater is generally interpreted as significant heterogeneity.15 To explore possible causes of heterogeneity, both subgroup and meta-regression analyses were conducted. Publication bias was assessed by the trim-and-fill method and the Egger’s test.16 The significance of Egger’s test is set at 10% or less. The trim-and-fill method can infer the existence of unpublished hidden articles from a filled Funnel plot and correct the meta-analysis by imputing the presence of missing studies to yield an unbiased pooled estimate.17 The aforementioned statistical analyses were completed with the Stata software release 12 (StataCorp, College Station, TX, USA).
Results
Article selection
After a careful literature retrieval and stringent eligibility appraisal, eleven out of 43 initially retrieved articles were included and summarized in this systematic review and meta-analysis.9–11,18–25 The PRISMA flowchart schematizing the selection of qualified studies is presented in Figure 1. The total sample size was 4,446, including 2,074 breast cancer patients and 2,372 control subjects. Two studies conducted in different locations were reported by Menzel et al,21 and they were handled separately, leading to a total of 12 studies in the final analysis.
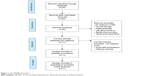  | Figure 1 The PRISMA flowchart. |
Study characteristics
Table 1 shows the baseline characteristics of all included studies. Out of 12 independent studies, four were performed in Caucasians, three in Asians, and five in mixed-ethnic populations. Two studies were designed longitudinally and ten cross-sectionally. Of the 12 studies, six studies had controls collected from hospitals and six from populations. The RFLP genotyping method was adopted in seven studies. Breast cancer patients were slightly older than control subjects (54.95 vs 50.94 years, P=0.070). Mean BMI levels were comparable between the two groups (P>0.999).
Overall estimates
As the study by Mandelblatt et al25 provided only ε4 positive/negative genotypes, overall allele comparisons were based on the remaining eleven studies (Figure 2). In overall analyses, using the most common ε3 allele as reference, the ε2 (OR =0.87, 95% CI =0.72–1.05, P=0.154, I2=0.0%) and ε4 (OR =1.07, 95% CI =0.80–1.42, P=0.654, I2=71.8%) alleles were found not to be significantly associated with breast cancer risk in a random-effects model using the DerSimonian and Laird method. The probability of publication bias was low according to the Egger’s test (for ε2 vs ε3, P=0.270, and for ε4 vs ε3, P=0.186); however, in the trim-and-fill method, three missing studies were filled for the comparison of alleles ε4 with ε3, in order to make the filled funnel plot symmetrical (Figure 3). After taking these three studies into consideration, the odds of breast cancer conferred by ε4 allele was 0.90 (95% CI =0.68–1.20, P=0.487), with significant heterogeneity (I2=75.4%).
  | Figure 2 Forest plots of APOE gene ε2 and ε4 alleles relative to ε3 allele in overall analyses. |
  | Figure 3 Filled funnel plots of APOE gene ε2 and ε4 alleles relative to ε3 allele in overall analyses. |
In addition, for the comparison of ε4 positive with ε4 negative genotypes, the odds of having breast cancer was 1.05 (95% CI =0.75–1.11, P=0.782) and heterogeneity was not significant (I2=46.1%) (Figure S1).
Stratified estimates
To examine whether effect estimates were homogeneous in subgroups by race, study design, source of controls, genotyping method, and sample size, as well as whether heterogeneity is improved, a set of subgroup analyses were carried out in a random-effects model using the DerSimonian and Laird method (Table 2). By race, no significance was noted for the comparison of alleles ε2 with ε3 in Caucasians, Asians, and mixed-ethnic populations, while the comparison of alleles ε4 with ε3 was significant in Asians only (OR =1.58, 95% CI =1.17–6.32, P=0.003) without heterogeneity (I2=12.1%). Grouping studies by study design and source of controls revealed only marginal significance for the comparison of alleles ε4 with ε3 in hospital-based studies (OR =1.33, 95% CI =0.98–1.79, P=0.065, I2=30.2%). Similarly, studies with the RFLP genotyping method showed a significant effect estimate of 1.27 (95% CI =1.01–1.61, P=0.045, I2=34.3%) for the comparison of alleles ε4 with ε3. In contrast, the presence of ε2 was found to be significantly associated with breast cancer risk only in small studies (total sample size <500) (OR =0.73, 95% CI =0.54–1.00, P=0.052) without heterogeneity (I2=0.0%).
Meta-regression analysis
Two steps were taken by modeling potential confounders in a meta-regression analysis. First, potential confounders including age, BMI, race, study design, source of controls, genotyping method, and sample size were modeled one at a time, and unfortunately there was no indication of significance in all regression models. Second, age, BMI, race, study design, source of controls, genotyping method, and sample size were modeled altogether and still no significant confounding effects were observed (all P>0.05).
Discussion
This is an updated systematic review and meta-analysis to assess the susceptibility of APOE gene ε2/ε3/ε4 polymorphism to breast cancer on the basis of eleven articles and 4,446 study subjects. Consistent with the meta-analytical results by Saadat,12 we in a larger sample size observed a significant association between APOE gene ε4 allele and breast cancer risk in Asians. In addition, our findings in subgroup analyses suggest that source of controls, genotyping platform, and sample size might be potential causes of heterogeneity.
There were experimental data reporting a high expression level of APOE gene in many forms of cancer, including breast cancer.11,26 Elevated APOE level was postulated to trigger tumor growth by providing lipid substrates for tumor cells.27 It was reported that different APOE isoforms affected tumor growth and proliferation at different levels, with ε2 allele having a largest protective effect, ε3 allele the moderate, and ε4 allele the least.28 Consistent with this tendency, in overall analyses, we observed a reduced risk of breast cancer for ε2 allele and an increased risk for ε4 allele with reference to ε3 allele; yet no statistical significance was detected. However, after restricting the analysis to populations of Asian descent, the presence of ε4 allele remarkably rendered the study subjects 58% more likely to develop breast cancer, suggesting a race-specific role of APOE gene in breast carcinogenesis. Besides genetic makeup, lifestyle backgrounds such as physical activity and dietary habits could also account for this race-specific divergence. Due to the limitation of unavailable lifestyle data, there is always a possibility of residual confounding.
Heterogeneity is a potential issue when interpreting the results of all meta-analyses in medical literature.29 In general, a meta-analysis can at least in part confirm the involvement of a genetic variant if heterogeneity is sufficiently recognized and taken into account. As revealed by our subgroup analyses, source of controls, genotyping platform, and sample size might be potential causes of heterogeneity. Taking source of controls as an example, it is widely believed that there is poor comparability between cases and controls in hospital-based studies given a regional specialty for the disease under study and differential hospitalization rates between cases and controls.30 In contrast, study subjects drawn from a community or a general population might be representative of a true population. In view of this divergence, the results from population-based studies might be convincing. Moreover, sometimes genotyping errors can induce biases in frequency estimates for APOE gene ε2/ε3/ε4 polymorphism. For example, partial digestion of PCR product in RFLP genotyping method may introduce a misclassification bias, and it is essential to have RFLP images read independently by two persons. Further we know that individual studies with small sample sizes may have not sufficiently statistical power to detect a small effect and yield a fluctuated estimate, and often wide CIs may indicate lack of precision. On the basis of these observations, it is high time to confirm the findings of this systematic review and meta-analysis in more population-based, larger association studies with validated genotyping methods.
This systematic review and meta-analysis is limited due to the following aspects. First, only articles in English language are included, and a selection bias cannot be excluded, although no observable publication bias was recorded by the Egger’s test. Second, only one polymorphism in APOE gene is analyzed and other polymorphisms in APOE gene and other susceptibility genes are not considered in this study, leading to the possibility that the potential role of APOE gene ε2/ε3/ε4 polymorphism is diluted or masked by other gene–gene and gene–environment interactions. Third, this systematic review and meta-analysis is not based on individual participant data and, therefore, adjusted effect-size estimates cannot be derived. Fourth, because mainly populations from Asia and Western countries were available, the present data did not allow us to extrapolate our findings to other racial or ethnic groups.
In conclusion, we in this updated systematic review and meta-analysis observed a significant association between APOE gene ε4 allele and breast cancer risk in Asians. Moreover, in our subgroup analyses, source of controls, genotyping platform, and sample size might be potential causes of heterogeneity. Further epidemiologic and functional validation is necessitated to provide a convincing effect estimate.
Acknowledgments
This study was supported by the Qiqihar Medical University Inner Project (QY2016B-06) and the Science and Technology Planning Project of Qiqihar City (SFGG-201205).
Disclosure
The authors report no conflicts of interest in this work.
References
Maxwell KN, Nathanson KL. Common breast cancer risk variants in the post-COGS era: a comprehensive review. Breast Cancer Res. 2013;15(6):212. | ||
Lee YH, Kim JH, Song GG. Genome-wide pathway analysis of breast cancer. Tumour Biol. 2014;35(8):7699–7705. | ||
Scollen S, Luccarini C, Baynes C, et al. TGF-beta signaling pathway and breast cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2011;20(6):1112–1119. | ||
Chen YC, Pohl G, Wang TL, et al. Apolipoprotein E is required for cell proliferation and survival in ovarian cancer. Cancer Res. 2005;65(1):331–337. | ||
Wu J, Yin RX, Guo T, et al. Gender-specific association between the cytoplasmic poly(A) binding protein 4 rs4660293 single nucleotide polymorphism and serum lipid levels. Mol Med Rep. 2015;12(3):3476–3486. | ||
Utermann G, Langenbeck U, Beisiegel U, Weber W. Genetics of the apolipoprotein E system in man. Am J Hum Genet. 1980;32(3):339–347. | ||
Gotoh N, Kuroiwa S, Kikuchi T, et al. Apolipoprotein E polymorphisms in Japanese patients with polypoidal choroidal vasculopathy and exudative age-related macular degeneration. Am J Ophthalmol. 2004;138(4):567–573. | ||
Weisgraber KH, Rall SC Jr, Innerarity TL, Mahley RW, Kuusi T, Ehnholm C. A novel electrophoretic variant of human apolipoprotein E. Identification and characterization of apolipoprotein E1. J Clin Invest. 1984;73(4):1024–1033. | ||
Cibeira GH, Giacomazzi J, Aguiar E, et al. Apolipoprotein E genetic polymorphism, serum lipoprotein levels and breast cancer risk: a case-control study. Mol Clin Oncol. 2014;2(6):1009–1015. | ||
Porrata-Doria T, Matta JL, Acevedo SF. Apolipoprotein E allelic frequency altered in women with early-onset breast cancer. Breast Cancer (Auckl). 2010;4:43–48. | ||
Surekha D, Vishnupriya S, Sailaja K, Nageswara R, Raghunadharao D. Influence of apolipoprotein E gene polymorphism on the risk for breast cancer. Int J Hum Genet. 2008;8(3):277–282. | ||
Saadat M. Apolipoprotein E (APOE) polymorphisms and susceptibility to breast cancer: a meta-analysis. Cancer Res Treat. 2012;44(2):121–126. | ||
Anand R, Prakash SS, Veeramanikandan R, Kirubakaran R. Association between apolipoprotein E genotype and cancer susceptibility: a meta-analysis. J Cancer Res Clin Oncol. 2014;140(7):1075–1085. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
Bowden J, Tierney JF, Copas AJ, Burdett S. Quantifying, displaying and accounting for heterogeneity in the meta-analysis of RCTs using standard and generalised Q statistics. BMC Med Res Methodol. 2011;11:41. | ||
Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. | ||
Moysich KB, Freudenheim JL, Baker JA, et al. Apolipoprotein E genetic polymorphism, serum lipoproteins, and breast cancer risk. Mol Carcinog. 2000;27(1):2–9. | ||
Niemi M, Kervinen K, Kiviniemi H, et al. Apolipoprotein E phenotype, cholesterol and breast and prostate cancer. J Epidemiol Community Health. 2000;54(12):938–939. | ||
Yaylim I, Bozkurt N, Yilmaz H, Isbir T, Isik N, Arikan S. The apolipoprotein E epsilon 4 allele is not a risk factor for Turkish breast cancer patients. Cancer Genet Cytogenet. 2003;146(1):86–87. | ||
Menzel HJ, Sarmanova J, Soucek P, et al. Association of NQO1 polymorphism with spontaneous breast cancer in two independent populations. Br J Cancer. 2004;90(10):1989–1994. | ||
Chang NW, Chen DR, Wu CT, et al. Influences of apolipoprotein E polymorphism on the risk for breast cancer and HER2/neu status in Taiwan. Breast Cancer Res Treat. 2005;90(3):257–261. | ||
Chang SJ, Hou MF, Tsai SM, et al. Association between the apolipoprotein E genotypes and breast cancer patients in Taiwanese. Breast Cancer Res Treat. 2006;98(1):109–113. | ||
McDonald BC, Conroy SK, Smith DJ, West JD, Saykin AJ. Frontal gray matter reduction after breast cancer chemotherapy and association with executive symptoms: a replication and extension study. Brain Behav Immun. 2013;30(Suppl):S117–S125. | ||
Mandelblatt JS, Stern RA, Luta G, et al. Cognitive impairment in older patients with breast cancer before systemic therapy: is there an interaction between cancer and comorbidity? J Clin Oncol. 2014;32(18):1909–1918. | ||
Niemi M, Hakkinen T, Karttunen TJ, et al. Apolipoprotein E and colon cancer. Expression in normal and malignant human intestine and effect on cultured human colonic adenocarcinoma cells. Eur J Intern Med. 2002;13(1):37–43. | ||
Rubinsztein DC, Hanlon CS, Irving RM, et al. Apo E genotypes in multiple sclerosis, Parkinson’s disease, schwannomas and late-onset Alzheimer’s disease. Mol Cell Probes. 1994;8(6):519–525. | ||
Miyata M, Smith JD. Apolipoprotein E allele-specific antioxidant activity and effects on cytotoxicity by oxidative insults and beta-amyloid peptides. Nat Genet. 1996;14(1):55–61. | ||
Yan S, Jiao X, Li K, Li W, Zou H. The impact of IGF-1R expression on the outcomes of patients with breast cancer: a meta-analysis. Onco Targets Ther. 2015;8:279–287. | ||
Niu W. The relationship between natriuretic peptide precursor a gene T2238C polymorphism and hypertension: a meta-analysis. Int J Hypertens. 2011;2011:653698. |
Supplementary material
References
Moysich KB, Freudenheim JL, Baker JA, et al. Apolipoprotein E genetic polymorphism, serum lipoproteins, and breast cancer risk. Mol Carcinog. 2000;27(1):2–9. | ||
Yaylim I, Bozkurt N, Yilmaz H, Isbir T, Isik N, Arikan S. The apolipoprotein E epsilon 4 allele is not a risk factor for Turkish breast cancer patients. Cancer Genet Cytogenet. 2003;146(1):86–87. | ||
Surekha D, Vishnupriya S, Sailaja K, Nageswara R, Raghunadharao D. Influence of Apolipoprotein E Gene Polymorphism on the Risk for Breast Cancer. Int J Hum Genet. 2008;8(3):277–282. | ||
Porrata-Doria T, Matta JL, Acevedo SF. Apolipoprotein E Allelic Frequency Altered in Women with Early-onset Breast Cancer. Breast Cancer (Auckl). 2010;4:43–48. | ||
Cibeira GH, Giacomazzi J, Aguiar E, et al. Apolipoprotein E genetic polymorphism, serum lipoprotein levels and breast cancer risk: A case-control study. Mol Clin Oncol. 2014;2(6):1009–1015. | ||
Mandelblatt JS, Stern RA, Luta G, et al. Cognitive impairment in older patients with breast cancer before systemic therapy: is there an interaction between cancer and comorbidity? J Clin Oncol. 2014;32(18):1909–1918. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



