Back to Journals » Drug Design, Development and Therapy » Volume 9
The mazEF toxin–antitoxin system as an attractive target in clinical isolates of Enterococcus faecium and Enterococcus faecalis
Authors Soheili S, Ghafourian S, Sekawi Z, Neela VK, Sadeghifard N, Taherikalani M, Khosravi A, Ramli R, Hamat RA
Received 9 November 2014
Accepted for publication 10 December 2014
Published 8 May 2015 Volume 2015:9 Pages 2553—2561
DOI https://doi.org/10.2147/DDDT.S77263
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Shu-Feng Zhou
Sara Soheili,1 Sobhan Ghafourian,2 Zamberi Sekawi,1 Vasantha Kumari Neela,1 Nourkhoda Sadeghifard,2 Morovat Taherikalani,2 Afra Khosravi,2 Ramliza Ramli,3 Rukman Awang Hamat1
1Department of Medical Microbiology and Parasitology, Faculty of Medicine and Health sciences, Universiti Putra Malaysia, Serdang, Malaysia; 2Clinical Microbiology Research Center, Ilam University of Medical Sciences, Ilam, Iran; 3Department of Medical Microbiology and Immunology, Faculty of Medicine, Universiti Kebangsaan Malaysia Medical Centre, Jalan Yaakob Latif, Bandar Tun Razak, Kuala Lumpur, Malaysia
Abstract: The toxin–antitoxin (TA) system is a regulatory system where two sets of genes encode the toxin and its corresponding antitoxin. In this study, the prevalence of TA systems in independently isolated clinical isolates of Enterococcus faecium and Enterococcus faecalis was determined, the dominant TA system was identified, different virulence genes in E. faecium and E. faecalis were surveyed, the level of expression of the virulence and TA genes in normal and stress conditions was determined, and finally their associations with the TA genes were defined. Remarkably, the analysis demonstrated higBA and mazEF in all clinical isolates, and their locations were on chromosomes and plasmids, respectively. On the other hand, a quantitative analysis of TA and virulence genes revealed that the expression level in both genes is different under normal and stress conditions. The results obtained by anti-mazF peptide nucleic acids demonstrated that the expression level of virulence genes had decreased. These findings demonstrate an association between TA systems and virulence factors. The mazEF on the plasmids and the higBA TA genes on the chromosomes of all E. faecium and E. faecalis strains were dominant. Additionally, there was a decrease in the expression of virulence genes in the presence of anti-mazF peptide nucleic acids. Therefore, it is suggested that mazEF TA systems are potent and sensitive targets in all E. faecium and E. faecalis strains.
Keywords: Enterococcus faecium, Enterococcus faecalis, toxin–antitoxin system, virulence genes, peptide nucleic acids
Introduction
Toxin–antitoxin (TA) systems, first described in the mid-1980s, are regulatory loci that encode a toxin and its corresponding antitoxin. The TA system could be RNA or a protein, but in all TA systems reported to date, the antitoxin has been found to be unstable while the toxin is stable.1 TA loci are often transferred by horizontal transformation and are more often associated with pathogenic bacteria; most of them have been found on plasmids containing antibiotic resistance,2 and virulence genes may also be harbored on TA plasmids. The use of toxins has been proposed as a new approach for antimicrobial therapy in pathogenic bacteria.3
The majority of clinical enterococcal infections are caused by Enterococcus faecalis and Enterococcus faecium. E. faecalis is considered to be more virulent; however, E. faecium is more likely to be antibiotic resistant.4 Twenty years ago, only 10% of the nosocomial enterococcal infections were caused by E. faecium.4 Now, approximately 40% of the enterococcal nosocomial infections worldwide are caused by E. faecium.5 This ratio changed in favor of E. faecium in the United States during the late 1990s and in Europe around the year 2000.6 In the last two decades, the emergence of enterococci as an important nosocomial pathogen has been increasingly documented. Unfortunately, the pathogenesis of enterococcal infections is only partly understood. However, several adhesins, hemolysin, hyaluronidase, aggregation substances, gelatinase, and genes encoding pili are now considered possible virulence factors.7 So far, at least 22 different genes, collectively called fms (E. faecium surface protein-encoding genes) are considered putative virulence factors in Enterococcus spp. Virulence factors encoded by acmfm (fms8), hyl, espfm, sgrA, and ecbA are most strongly associated with clinical lineages in E. faecium.8 Antisense therapies, which are sequence dependent, silence a specific gene. The antisense components are analogs of messenger (m)RNA; therefore, this technology is involved in the inhibition of gene expression. Many techniques are available for antisense therapy that use different RNA analogs, such as phosphorodiamidate morpholino oligomers, locked nucleic acids, and peptide nucleic acids (PNA). Among these, the properties of PNA make them particularly appropriate for antisense therapy in bacteria. This technique is applied for molecular bioengineering, therapeutic methods, and antibiotics.9–11 The structure of PNAs is similar to that of DNA or RNA, except that the nucleobases are changed to a pseudopeptide12 following the Watson and Crick base-pairing rule; however, PNAs can bind DNA and RNA.13
The TA system could be a potent target for antibiotic therapy. In theory, the activation of a toxin or the inhibition of an antitoxin is an attractive strategy for antimicrobial therapy.14,15 Amitai et al16 demonstrated that 5% of bacterial cells were viable and 95% were killed after toxin activation because the increased toxin could not be neutralized by the antitoxin. However, when coexpressing mazE (antitoxin) and neutralizing mazF (toxin), 85% of the cells were viable because the toxin was neutralized and inhibited by the antitoxin.16 Nonetheless, the most important step for potency of the TA system, as a target, is to identify a TA system that is prevalent in all pathogenic clinical strains and to determine its functionality. While the analysis of a TA system can be instructive, until now, there has been no information available on the prevalence and identity of TA systems in pathogenic E. faecium and E. faecalis. Therefore, it is necessary to study a TA system that is prevalent and transcribed in all clinical pathogenic E. faecium and E. faecalis strains and to evaluate the TA system as a potent target in E. faecium and E. faecalis.
Materials and methods
Ethics statement
The ethical committee members of the Universiti Putra Malaysia (Selangor, Serdang, Malaysia) approved (JMMP-July [10] 02) the collection of wound samples for the identification of Enterococcus spp. from patients; all patients and the parents or guardians of children in this study provided their written consent. The ethical committee of the Universiti Putra Malaysia specifically approved this study.
Bacterial isolates
A total of 79 clinical isolates of E. faecalis and E. faecium were identified during the period of May 2009–March 2010 from a tertiary teaching hospital. Of these Enterococcus isolates, 29 were E. faecium and 50 isolates were E. faecalis. The isolates were collected from urine, blood, pus, vaginal, and sterile body fluids.
Evaluation of different virulence genes
All of the isolates were subjected to the amplified pilA, pilB, hylA, ecbA, scm, fms8, efaAfm, and sgrA genes using the specific primers that were designed.
Evaluation of the TA systems
All the clinical isolates of E. faecium and E. faecalis were subjected to polymerase chain reaction (PCR) using total chromosomal or plasmid DNA. Oligodeoxy nucleotide primer pairs were designed for specific genes using sequences obtained from GenBank (European Nucleotide Archive: http://www.ebi.ac.uk/ena/). Primers were synthesized to amplify the mazEF, relBE, and higBA TA genes.
Plasmid transformation
For confirmation that the TA genes were harbored by plasmids, a transformation was performed. The plasmid-free strain Staphylococcus aureus RN4220 was used as a transforming host. Plasmid DNA was extracted from the clinical isolates using a plasmid extraction kit (Thermo Fisher Scientific, Waltham, MA, USA) and was then fractionated into agarose gel by electrophoresis. Each plasmid-specific band was purified from the agarose gel using the QIAEX II® Gel Extraction Kit. The isolated plasmids were then transformed into the S. aureus RN4220 competent cells. For confirmation that the plasmids harbored the TA genes, different purified plasmids were subjected to PCR.
Sequence analysis
The PCR products of the virulence and TA genes were purified from gel agarose, and the purified products were sequenced by Sigma-Aldrich Co. (St Louis, MO, USA). The results of the DNA sequencing were run in the Chromas Lite program to analyze their similarity to the sequenced gene in the GenBank library.
Stress induction
The stress on the E. faecium and E. faecalis strains was induced by heat stress aqua: E. faecium and E. faecalis were shifted from 37°C to 42.5°C for 30 minutes, as described by Anderson et al.17 RNA was extracted from the bacteria in normal and stress conditions, and the level of expression of the TA systems and virulence genes was evaluated by real-time quantitative (RT-q)PCR.
RT-qPCR
The specific primers and TaqMan probes for the TA genes, the virulence genes, and the 23S ribosomal (r)RNA (a reference gene) were all designed using the GenScript software and were synthesized by Sigma-Aldrich Co. The ability of the primers for each specific gene to amplify the appropriate amplicon length was evaluated using 4 μL of complementary (c)DNA in a total volume of 20 μL per reaction in a Mastercycler® realplex 2 (Eppendorf, Hamburg, Germany) with the TaqMan® Fast Universal PCR Master Mix. The thermocycling conditions consisted of an initial denaturation for 15 minutes at 95°C, followed by 40 cycles of 95°C for 15 seconds, 60°C for 40 seconds and 68°C for 20 seconds. The RT-qPCR results were then analyzed quantitatively to estimate the level of TA and the virulence transcription factors in E. faecium and E. faecalis by calculating the number of mRNAs that were produced.
PNA design
The mazF toxin was targeted for the antisense PNA therapy. Specific PNA was designed for the mazF toxin. The mazF (+10 start codon) PNA sequence was designed and synthesized by Promega Corporation (Fitchburg, WI, USA). The PNA structure was mazF:KFFKFFKFFK-eg-ATGATTAGAC-CONH2.
Antisense PNA therapy of E. faecium and E. faecalis containing the mazEF TA locus plasmid
A 0.5 McFarland standard of clinical isolates of E. faecalis (three isolates) and E. faecium (two isolates) containing the plasmid mazEF TA locus and all the virulence genes were prepared. PNA of different concentrations (1–10 μmol) in each well of a Costar microplate, in combination with 0.5 McFarland standards of E. faecalis and E. faecium, were prepared. The negative control was Mueller Hinton broth, and the positive control was E. faecium without PNA. The Costar microplate was incubated at 35°C. The effects of PNA at different concentrations (1–10 μmol) against E. faecalis and E. faecium were examined. RT-qPCR was performed to check the expression level of mazF and the virulence genes.
Results and discussion
The mazEF and higBA TA genes are prevalent in all clinical isolates of E. faecium and E. faecalis
In the current study, both plasmid and chromosomal DNA were subjected to amplification by PCR to identify the prevalence of different TA genes and their location on the plasmid or chromosome. For this purpose, after extraction of the chromosomal and plasmid DNA from the E. faecalis and E. faecium strains, the specific primers were used to amplify the mazEF, relBE, and higBA TA genes. All chosen TA systems belong to a type II TA system; thus, both the toxin and antitoxin are proteins. For accuracy of the PCR results, the toxin and antitoxin were separately subjected to amplification. The results were confirmed with the presence of both the toxin and antitoxin genes. The results of the PCR analysis demonstrated that the TA genes were more prevalent on the plasmid than on the chromosome; the exception was the higBA TA system. The prevalence of the TA systems is displayed in Figure 1.
  | Figure 1 Prevalence of the different TA loci among Enterococcus faecium and Enterococcus faecalis clinical isolates. |
The mazEF TA system was found on the plasmids of all E. faecium and E. faecalis strains, but it was present in only 59.4% (number [n]=43/79) of the chromosomal DNA samples. It could be concluded that mazEF was likely harbored by the plasmid; this can be confirmed by plasmid transformation. Evaluation of the relBE TA system showed that 59.5% (n=47/79) of the E. faecium and E. faecalis strains were positive for the relBE TA system on the plasmid. However, lower prevalence rates (19% [n=15/79]) were observed in the chromosomal DNA. When evaluating E. faecium strains for the presence of higBA TA loci, 39.2% (n=31/79) of the plasmid DNA and 100% (n=79/79) of the chromosomal DNA possessed the higBA TA system.
Therefore, the mazEF and higBA TA loci had a 100% prevalence among E. faecium and E. faecalis. Thus, the mazEF and higBA TA systems are putative targets that are prevalent in all E. faecium and E. faecalis strains.
After using the plasmid extraction kit, the plasmid was subjected to adenosine triphosphate-dependent DNase, which digested the linear DNA, but was not effective against double-strand super-coiled DNA. This confirmed that there was no contamination in the chromosomal DNA. The confirmation was made through plasmid transformation into the S. aureus RN4220 free plasmid as well. The next section shows that the mazEF TA locus is harbored by a 20 kb plasmid.
The mazEF TA system is harbored by a plasmid while the possible location of relBE is on the chromosome
To confirm that the TA locus was harbored by a plasmid and not contaminated by the chromosomal DNA, a transformation was performed. After extraction of the plasmid from E. faecium using the plasmid extraction kit, the plasmids were fractionated on 1% agarose gel. Then, each plasmid was purified using the QIAEX II® Gel Extraction Kit, which is able to purify DNA fragments between 100 bp and 50 kb in length. As shown in Figure 2, most E. faecium and E. faecalis strains contain two plasmids, while some of the strains contain three plasmids.
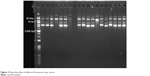  | Figure 2 Plasmid profiles of different Enterococcus spp. strains. |
After purification of each plasmid, they were subjected to PCR to probe for the mazEF and higBA TA loci. The 20 kb plasmid had a positive result for mazEF, while negative results were obtained for higBA in all the purified plasmids. Thus, the 20 kb plasmid was transformed to the S. aureus RN4220 plasmid-free strain by electroporation. Plasmid extraction was performed on the S. aureus RN4220 containing the mazEF TA system plasmid (Figure 3). PCR was performed to confirm the presence of the mazEF TA locus on the 20 kb plasmid. These results confirmed that the mazEF TA system is harbored by the plasmid. The PCR results of the plasmid confirmed the presence of mazEF and the absence of higBA. Therefore, the mazEF TA locus is harbored by the plasmid, and the bacterial chromosome potentially possesses the higBA TA locus.
pilB, fms8, efaAfm, and sgrA are the most prevalent virulence genes in E. faecium and E. faecalis
A total of 79 E. faecalis (50 isolates) and E. faecium (29 isolates) isolates from clinical infections were analyzed for the presence of different virulence genes including pilA, pilB, hylA, ecbA, scm, fms8, efaAfm, and sgrA. The analysis showed that the prevalence of the different virulence genes in Enterococcus spp. ranged from 35.4% to 100%. pilB, fms8, efaAfm, and sgrA were the most prevalent virulence genes and were observed in all 79 isolates. The second most prevalent virulence gene, scm, was found in 92.4% of the isolates (n=73/79). ecbA was determined to be the third most prevalent virulence gene with a frequency of 81% (n=64/79). pilA had a prevalence of 73.4% (n=58/79) in the clinical isolates of Enterococcus spp. The lowest prevalence was reported in hyl with a frequency of 35.4% (n=28/79). The prevalence rates of the different virulence genes are shown in Figure 4. The analysis also showed that all the selected virulence genes were found in E. faecium and that hyl was predominant with a prevalence rate of 82.7%. In E. faecalis, all the virulence genes with different prevalence rates were observed, as shown in Table 1. This is the first report of the prevalence of selected virulence genes in E. faecalis, which demonstrates the possibility of horizontal transformation between E. faecium and E. faecalis.
  | Table 1 Distribution of the different virulence factors in E. faecium and E. faecalis |
Functionality of TA loci based on an evaluation of the quantity of toxin and antitoxin
To assess the level of expression of mazE and mazF, relB and relE, and higB and higA, RT-qPCR was performed. The level of expression of each TA gene in stressed cultures was evaluated relative to the control cells. The experiment was compared with that under normal conditions. The results under normal conditions had almost the same quantity of toxin and antitoxin, as indicated in Figure 5. The results of the heat stress showed an increase in toxin levels and a decrease in antitoxin levels. The reason for this increase in toxin and decrease in antitoxin could be explained by the degradation of the antitoxin. Greater transcription occurred because the TA system had the same promoter, and the antitoxin was unstable and was degraded by a protease. Therefore, the amount of toxin increased. However, the analysis by RT-qPCR showed that stress could increase the expression of toxin and decrease antitoxin expression.
In 2009, Ramage et al18 demonstrated the functionality of the TA system under stress using RT-qPCR. They identified 88 different TA systems in Mycobacterium tuberculosis.18 They then induced stress by hypoxia and phagocytosis by macrophages and observed the functionality of four different TA systems. Antitoxin degradation led to increased toxin transcription. Thus, the results indicated an increase in toxin and antitoxin mRNA. However, the antitoxin, which is unstable, was targeted by a protease, leading to an increase in free toxin.19 An increase in toxin expression and a decrease in antitoxin expression were also observed in the present study.
A study by Wang and Wood20 evaluated the mqsRA TA system (mqsR is the antitoxin, and mqsA is the toxin) under oxidative stress and tested its effect on the rpoS gene, which is responsible for controlling 500 genes in Escherichia coli, including biofilm formation. Their findings revealed that the antitoxin (mqsR) caused a decrease in the expression of the rpoS gene under normal conditions, leaving the E. coli unable to form biofilms. Under oxidative stress conditions, the antitoxin was degraded, and the rpoS gene was induced, resulting in biofilm formation.20 Therefore, the results of the current study are consistent with those of Wang and Wood20 and indicate that stress can cause antitoxin degradation.
Quantity evaluation of different virulence genes in normal and stress conditions
The expression levels of the virulence factors of the E. faecalis and E. faecium clinical strains in normal conditions, and their response to heat stress, were determined using RT-qPCR (Figure 6). As shown in Figure 6, the expressions of all the virulence genes examined in normal conditions were functional, and with heat stress, the expression level of all the virulence genes showed an increase. Therefore, the pathogenicity of E. faecalis and E. faecium might have increased. Only one study has demonstrated the role of stress in the expression level of virulence genes in E. faecalis. There are no studies that explain the expression level of virulence genes under normal and stress conditions in E. faecium. In 2010, Lenz et al21 demonstrated that sublethal stress could cause an increase in the pathogenicity of E. faecalis by increasing the expression level of virulence genes. E. faecalis were exposed to sublethal osmotic, heat, high hydrostatic pressure, acid, and bile salt stress. Almost the same results were obtained, which indicated that stress causes more pathogenicity in E. faecalis. The results obtained in the current study were consistent with the findings by Lenz et al21 in that there was an increase in the expression level of all the virulence genes in E. faecalis and E. faecium in the 79 clinical isolates. In the next section, the relationship between virulence genes and TA systems will be discussed.
The mazEF TA system has a direct role in the regulation of virulence genes
As the mazEF TA locus was prevalent in all clinical strains of E. faecalis and E. faecium, it was targeted for silencing by anti-mazF PNA. Antisense therapy causes the silencing of a specific gene. This technique is applied in molecular bioengineering, therapeutic methods, and antibiotics.9–11 PNA is an analog of nucleotide sequences. It has been found to be the best complementary strand of DNA and RNA.12 PNA has many properties that make it unique, such as its stability against proteases and nucleases, and its stability in high concentrations of salt and at high temperatures.22 Another important property of PNA for efficiency is its size. As normal PNA is larger than an antibiotic, it must be designed to be smaller to pass through the cell. It is also important that the synthesized PNA is efficient against the target gene. The mazF (antitoxin) is approximately 171 bp in size. The PNA was designed based on the start codon and the ten nucleotides before the start codon region of the mazF gene. The size of the PNA was 10 mer (shorter than common PNAs). Then, the anti-mazF PNA was evaluated against E. faecium and E. faecalis strains containing the mazF gene at different concentrations in Mueller Hinton broth. The results were evaluated by RT-qPCR for determination of the specificity of the anti-mazF PNA and its association with the expression level of virulence genes.
As shown in Figure 7, with increases of PNA, the amount of mazF to be silenced and also the quantity of virulence genes showed a decrease. Therefore, these findings show the direct role of the TA system on the pathogenicity of E. faecium and E. faecalis. PNA antisense therapy is a good technique for antisense therapy. The results obtained by Good et al23 showed 2 μM anti-acpP PNA SP4 and no viable bacterial cells. Goh et al24 showed the RNA silencing aspect to bacterial growth. They revealed that by reducing the mRNA PNA, a 50% reduction was observed in the cell growth of E. coli.24
Some limitations of this study were the high cost of performing the PNA antisense therapy on a large scale and the higBA TA system.
Conclusion
This study describes and validates a novel target, mazF, which is found in clinical isolates of E. faecium and E. faecalis strains. First, the mazEF TA locus was shown to be present in all E. faecium strains. We showed that the mazEF and higBA loci are ubiquitous in a collection of E. faecium and E. faecalis clinical isolates. The expression and function of the mazEF, relBE, and higBA TA loci in E. faecium and E. faecalis strains was confirmed by RT-qPCR. Therefore, the TA system appears to be reliably expressed in those strains that pose a particular challenge in medicine. Second, inactivation of the mazF toxin using an anti-mazF PNA selectively decreased the expression of the virulence genes. Therefore, inactivation of the mazF toxin has been validated as a novel antimicrobial strategy. This research provides a first step in the introduction of the mazEF TA system as a sensitive target; however, further studies are needed to test the effectiveness of mazF in vivo. Furthermore, the occurrence and potential for activation of the mazEF TA system in other pathogenic bacteria remains to be investigated.
Disclosure
The authors report no conflicts of interest in this work.
References
Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfih entities? PLoS Genet. 2009;5(3):e1000437. | ||
Mine N, Guglielmini J, Wilbaux M, Van Melderen L. The decay of the chromosomally encoded ccdO157 toxin-antitoxin system in the Escherichia coli species. Genetics. 2009;181(4):1557–1566. | ||
Mutschler H, Meinhart A. ε/ζ systems: their role in resistance, virulence, and their potential for antibiotic development. J Mol Med (Berl). 2011; 89(12):1183–1194. | ||
Willems RJ, van Schaik W. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 2009; 4(9):1125–1135. | ||
van Schaik W, Willems RJ. Genome-based insights into the evolution of enterococci. Clin Microbiol Infect. 2010;16(6):527–532. | ||
Treitman AN, Yarnold PR, Warren J, Noskin GA. Emerging incidence of Enterococcus faecium among hospital isolates (1993 to 2002). J Clin Microbiol. 2005;43(1):462–463. | ||
Sava IG, Heikens E, Huebner J. Pathogenesis and immunity in enterococcal infections. Clin Microbiol Infect. 2010;16(6):533–540. | ||
Sillanpää J, Prakash VP, Nallapareddy SR, Murray BE. Distribution of genes encoding MSCRAMMs and Pili in clinical and natural populations of Enterococcus faecium. J Clin Microbiol. 2009;47(4):896–901. | ||
Lee LK, Roth CM. Antisense technology in molecular and cellular bioengineering. Curr Opin Biotechnol. 2003;14(5):505–511. | ||
Janson CG, Matthew JD. Peptide Nucleic Acids, Morpholinos and Related Antisense Biomolecules. New York, NY: Springer; 2006. | ||
Rasmussen LC, Sperling-Petersen HU, Mortensen KK. Hitting bacteria at the heart of the central dogma: sequence-specifi inhibition. Microb Cell Fact. 2007;6:24. | ||
Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254(5037):1497–1500. | ||
Jensen KK, Orum H, Nielsen PE, Nordén B. Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry. 1997;36(16):5072–5077. | ||
DeNap JC, Hergenrother PJ. Bacterial death comes full circle: targeting plasmid replication in drug-resistant bacteria. Org Biomol Chem. 2005;3(6):959–966. | ||
Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004;12(2):66–71. | ||
Amitai S, Yassin Y, Engelberg-Kulka H. MazF-mediated cell death in Escherichia coli: a point of no return. J Bacteriol. 2004;186(24): 8295–8300. | ||
Anderson KL, Roberts C, Disz T, et al. Characterization of the Staphylococcus aureus heat shock, cold shock stringent, and SOS responses and their effects on log-phase mRNA turnover. J Baceteriol. 2006;188(19):6739–6756. | ||
Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009; 5(12):e1000767. | ||
Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A. 2001;98(25):14328–14333. | ||
Wang X, Wood TK. Toxin-antitoxin systems inflence biofim and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77(16):5577–5583. | ||
Lenz CA, Hew Ferstl CM, Vogel RF. Sub-lethal stress effects on virulence gene expression in Enterococcus faecalis. Food Microbiol. 2010;27(3):317–326. | ||
Demidov VV, Potaman VN, Frank-Kamenetskii MD, et al. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol. 1994;48(6):1310–1313. | ||
Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. Bactericidal antisense effects of peptide-PNA conjugates. Nat Biotechnol. 2001; 19(4):360–364. | ||
Goh S, Boberek JM, Nakashima N, Stach J, Good L. Concurrent growth rate and transcript analyses reveal essential gene stringency in Escherichia coli. PLoS One. 2009;4(6):e6061. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





