Back to Journals » Therapeutics and Clinical Risk Management » Volume 14
The association between rs12901499 polymorphism in SMAD3 gene and risk of osteoarthritis: a meta-analysis
Authors Gao ST, Lv ZT , Sheng WB
Received 2 February 2018
Accepted for publication 14 March 2018
Published 15 May 2018 Volume 2018:14 Pages 929—936
DOI https://doi.org/10.2147/TCRM.S164409
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Deyun Wang
Shu-Tao Gao,1 Zheng-Tao Lv,2 Wei-Bin Sheng1
1Department of Spine Surgery, The First Affiliate Hospital, Xinjiang Medical University, Urumqi, Xinjiang, China; 2Department of Orthopedics, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei, China
Purpose: This study was conducted to assess and synthesize the current evidence on the association between rs12901499 polymorphism in SMAD3 gene and risk of osteoarthritis (OA).
Materials and methods: Four electronic databases, including PubMed, Embase, ISI Web of Science, and CENTRAL were systematically searched for potential studies. Summary odds ratio and corresponding 95% CI were calculated to evaluate the association. Risk of bias was assessed through the Newcastle–Ottawa Scale. Subgroups and sensitivity analyses were performed using the RevMan 5.3 software. Publication bias was evaluated by Egger’s and Begg’s tests. Power analysis was conducted using the Power and Sample Size Calculation program.
Results: Eight case–control studies containing 5,625 patients with OA and 5,600 healthy controls were obtained for the meta-analysis. After excluding cohorts with inadequate power, the pooled data supported that G allele carriers of rs12901499 had a significantly increased risk of OA (odds ratio 1.31, 95% CI: 1.21 to 1.43, P<0.00001). When stratified by OA site and ethnicity, the association remained statistically significant.
Conclusion: The combined results evidently supported that rs12901499 polymorphism in SMAD3 gene is significantly associated with OA vulnerability across both Caucasian and Asian populations.
Keywords: osteoarthritis, polymorphism, SMAD3, rs12901499, meta-analysis
Introduction
Osteoarthritis (OA), a disease of the joint as an organ, is one of the most common degenerative musculoskeletal disorders predominantly affecting synovial joints, such as knees, hips, spines, and hands.1 An individual joint could be involved, but in more cases, several joints are affected concomitantly. Disease progression of OA is typically slow, and it might take years or even decades to develop. Therefore, the dangerousness of OA always eludes the attention of patients until an advanced stage of this disorder. It is reported that ~240,000,000 elderly population suffers from OA around the world.2 With the prolongation of human lifespan, activity incapacity caused by knee OA alone is greater than any other medical conditions among population aged >65 years. The highly prevalent and attendant disability of OA has a formidable impact on both individuals and society.3,4 While, up to date, no effective treatment strategy for this common disease is still available. There is a great necessity to look for novel strategies to decrease the influence of this disturbing condition.
The underlying etiopathogenesis of OA remains largely indeterminate. Epidemiological studies have demonstrated that OA is driven by a host of factors, including age,5 gender,6 body mass index,7 as well as genetics.8 Involvement of genetic components in OA has attracted significant interest during the past 2 decades. In reality, researchers have observed familial aggregation phenomenon of OA for a long time.9 Evidence based on twin studies suggested an estimated heritability of 37%–65% of OA in different skeletal sites.10 Considerable single-nucleotide polymorphisms (SNPs) in candidates such as the estrogen-receptor,11,12 calmodulin,13,14 vitamin D receptor,15 ADAM12,16 and growth differentiation factor 5 genes17 have been reported to be linked to OA.
SMAD3 belongs to SMAD family, which consists of at least 8 different SMADs in human. It is an intracellular mediator molecule regulating the extracellular transforming growth factor beta (TGF-β) signals to transduce into nucleus. TGF-β/SMAD3 signaling is crucial to maintain the integrity of articular cartilage.18 SMAD3 gene-deficient mice are characterized with hypertrophic chondrocytes, which induce the progressive loss of cartilage accompanied by formation of osteophytes in joints.18 A clinical study led by Yao et al found that the SMAD3 gene mutations were associated with the pathogenesis of human OA.19
Albeit several reports have been published in an effort to demonstrate the correlation between rs12901499 polymorphism in SMAD3 gene and OA predisposition among different populations, it is quite regrettable that no convincing evidence is yet available, which might be owing to reasons such as limited sample size, inadequate statistical power, ethnicity specificity, heterogeneity of participants. Therefore, we turn to exploit a meta-analysis to synthesize the existing evidence on the association between rs12901499 polymorphism in SMAD3 gene and risk of OA.
Materials and methods
This systematic review and meta-analysis were conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines.20
Literature search strategy
Literature search of the study was conducted on PubMed, Embase, ISI Web of Science, and Cochrane Library (CENTRAL) 4 electronic databases, with all articles published previous to January 1, 2018. A combination of Medical Subject Headings (MeSH) together with free terms was applied to retrieve all the potentially eligible publications without language restriction. The following search string was employed: (Single Nucleotide Polymorphism or SNP or SNPs or “Polymorphism, Single Nucleotide” [Mesh]) and SMAD3 and (osteoarthritis or OA or osteoarthritides or osteoarthrosis or “Osteoarthritis” [Mesh]). Bibliographic lists of relevant reviews and full-text articles were also manually searched to capture more possible studies.
Inclusion and exclusion criteria
Studies meeting the following criteria were included for our meta-analysis: 1) studies examining the correlation between the rs12901499 polymorphism in SMAD3 and OA susceptibility that have been published; 2) case–control designed study; 3) studies with sufficient data to calculate the odds ratio (OR) with its 95% CI. Correspondingly, studies satisfying the following criteria were excluded: 1) animal study, review, expert opinion, conference abstract, case report, or case series; 2) data overlapping with previous publications; 3) studies failed to provide sufficient data to evaluate OR and the associated 95% CI.
Methodological quality assessment
Methodological quality of eligible studies was assessed independently by the first 2 reviewers using the Newcastle–Ottawa Scale (NOS) for observational studies.21 A “star system” has been developed in which a study is judged on 3 broad perspectives, including the selection of the study groups, the comparability of the groups, and the ascertainment of either the exposure or outcome of interest. Discrepancies between reviewers were settled through discussion until consensus was reached, a third reviewer could be consulted when necessary (W-BS).
Data extraction
Two independent investigators (S-TG and Z-TL) conducted the data extraction of all the included studies independently. A standardized data extraction list was utilized to collect information, including first author, year of the publication, country, sample size, ethnicity of enrolled subjects, site of OA, risk allele percentage of cases and controls, the calculated OR and 95% CI of individual cohort, and P-value for Hardy–Weinberg equilibrium (HWE) test of control subjects in each study. In the event of any discrepancy through the process, 2 reviewers reinvestigated the article together and discussed with each other until coming to an agreement.
Statistical analysis
To test the potential association between SMAD3 gene polymorphism (rs12901499) and risk of OA, the estimated allelic effect of G (the variant allele) versus A (the common allele) was indicated as OR and corresponding 95% CI using the allele frequency data extracted from included studies. Heterogeneity across included studies was estimated using a Q-test and the Higgins I2 test, where P>0.1 and I2<50% indicated acceptable heterogeneity. I2 values of 0%, 25%, 50%, and 75% were defined as no, low, moderate, and high heterogeneity, respectively. In our present study, we combined the OR of each study using a random-effect model because it accommodates diversity between studies and is preferable in the presence or anticipation of inter-study variances.22
Prior to the meta-analysis, power analysis was performed using the Power and Sample Size Calculation program (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize) to evaluate whether the selected studies could offer adequate power to detect the association between SMAD3 polymorphism and OA at a level of significance of 0.05. Studies that failed to provide adequate power (<0.50) were not included in the meta-analysis. Regardless of the heterogeneity detected between included studies, we performed subgroup-analysis by ethnicity and site of OA. The leave-one-out sensitivity analysis was conducted by removing each included study at a time and reevaluating the resulting effect on pooled results to test the robustness of summary risk estimate. Begg’s rank correlation test and Egger’s linear regression test using Stata version 12.0 (Stata Corp LP, College Station, TX, USA) were combined to assess the publication bias, which was considered present with P<0.05.23 Publication bias was also assessed by visually inspecting funnel plots, a visual asymmetry suggested possible publication bias. The forest and funnel plots were generated via RevMan 5.3 software (The Nordic Cochrane Centre, Copenhagen, Denmark).
Results
Literature search
The comprehensive literature retrieval of 4 electronic databases totally yielded 48 potentially relevant records comprising 16 from PubMed, 19 from Embase, 13 from ISI Web of Science, but none from CENTRAL. After the first scan, 20 duplicate records were deleted. Of the remaining 28 records, a further 18 citations were eliminated after title and abstract screening. Among the rest 10 articles, 2 unrelated were removed after assessment in the light of predefined inclusion and exclusion criteria. Ultimately, 8 studies24–31 were incorporated into the qualitative synthesis. The process of literature selection process is presented in Figure 1.
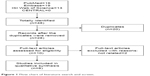  | Figure 1 Flow chart of literature search and screen. |
Main characteristics
The main characteristics of included studies are displayed in Table 1. A total of 8 case–control studies with 16 cohorts involving OA of the knee, hip, hand, spine, as well as temporomandibular joint (TMJ) were identified and included in the meta-analysis. All the included articles were published between 2010 and 2018. The sample size of each study ranged from 240 to 6,100. Six studies24,26,27,29–31 were performed among Asian population, and the other 225,28 across Caucasian population. The study by Valdes et al28 consisted of 5 independent studies involving 8 cohorts with knee or hip OA patients in case groups, and the study by Jiang et al24 comprised 2 cohorts with hand or knee OA patients. Liva et al study25 was about spine OA, and Xiao et al study29 was on TMJ OA. All these included cohorts conformed to HWE with the exception of Zhong et al.31 Of note, the statistical powers of these studies varied from 0.11 to 1.00, and several cohorts lacked adequate power (detailed data presented in Table 1). On the basis of the NOS, each study received no less than 5 stars for methodological quality assessment, as shown in Table 2.
  | Table 2 Quality assessment of included studies |
Meta-analysis and subgroup-analysis
After removing the cohorts with inadequate power, the pooled data suggested a significant correlation between rs12901499 polymorphism and risk of OA in the overall population with a modest heterogeneity (G versus A: OR 1.31, 95% CI: 1.21 to 1.43, P<0.00001; I2=29%; Figure 2). We performed subgroup-analysis by site of OA (knee, hip, hand, and TMJ) and ethnicity (Asian and Caucasian) to reappraise the association in different subgroups. Regarding OA site, subgroups of knee OA (OR 1.32, 95% CI: 1.17 to 1.48, P<0.00001; I2=35%), hip OA (OR 1.22, 95% CI: 1.08 to 1.37, P=0.001; I2=0), hand OA (OR 1.68, 95% CI: 1.21 to 2.33, P=0.002), and TMJ OA (OR 1.57, 95% CI: 1.09 to 2.26, P=0.01) all conformed to the association (Figure 3). In terms of ethnicity, there still existed a statistically significant association of rs12901499 polymorphism and OA in population of Asians (OR 1.47, 95% CI: 1.22 to 1.78, P<0.0001; I2=42%) and Caucasians (OR 1.24, 95% CI: 1.15 to 1.35, P<0.00001; I2=0), as shown in Figure 4.
Sensitive analysis and publication bias
Sensitivity analyses were tested by removing an individual dataset sequentially. But the pooled results did not change remarkably after the removal of any dataset, which suggested the stability and credibility of the overall estimate of risk (detailed data not shown). For publication bias investigation, there was an obvious symmetry presented in the funnel plot (Figure 5). Begg’s rank correlation test and Egger’s linear regression test also confirmed significant publication bias (Begg’s test: z=2.40, P=0.016; Egger’s test: t=8.02, P<0.001).
  | Figure 5 Funnel plot of rs12901499 in SMAD3 gene and risk of osteoarthritis. |
Discussion
Despite identification of the accurate risk factors for OA might greatly benefit earlier treatment in addition to preventive strategy, the precise etiology of OA remains unanswered. Researches of OA have focused on multiple areas and acknowledged the complexity of this disorder. Intrinsic factors in synergy with extrinsic factors are considered to be the potential etiology. As an intrinsic factor, SMAD3 gene has attracted a great deal of attention over the recent years. The association between rs12901499 polymorphism in SMAD3 gene and OA vulnerability has been documented in several studies with conflicting outcomes. To overcome the drawbacks of limited sample size and ethnicity specificity of an individual study, we designed the present meta-analysis to investigate the underlying correlation of rs12901499 and OA in populations from different ethnicities. The pooled data supported G allele carriers of rs12901499 had a significantly increased risk of OA (OR 1.31, 95% CI: 1.21 to 1.43, P<0.00001). Likewise, the association remained significant regardless of the site of OA or any ethnicity of the included populations, which suggested the convincing evidence that rs12901499 polymorphism in SMAD3 gene contributed to OA predisposition.
SMADs are a cluster of proteins sharing similar structure but having distinct roles in signaling.32 Of them, SMAD3 has been popularly investigated. A large body of evidence has established that changes in the normal structure of SMAD3 molecule implicated in numerous physiological and disease processes.33 SMAD3 gene is located in chromosome 15q22.33, a region, which has been reported to be linked with OA.34 The expression of SMAD3 gene plays a key role in the TGF-β pathway which is critical to TGF-β signal transduction.32,35 Reports have indicated that the TGF-β signaling promotes the metabolism of chondrocytes and assists in the maintenance of the normal phenotype of chondrocytes by regulating phosphorylation of SMAD3. In articular cartilage tissues, phosphorylated SMAD3 can protect chondrocyte phenotype from deleterious hypertrophy and production of catabolic enzymes.18 Li et al found that loss of SMAD3 could give rise to accelerated maturation of chondrocytes, the TGF-β signaling effects were as well decreased in SMAD3−/− chondrocytes.36 Van de Laar et al reported that mutations in SMAD3 gene could lead to serious aortic aneurysms and dissections accompanied by early-onset OA.37 All these investigations supported the importance of SMAD3 gene to OA, and targeting of SMAD3 may provide a novel strategy for the treatment of OA.
It should be pointed out that the study led by Valdes et al28 also involved a meta-analysis suggesting a significant association between rs12901499 polymorphism and OA susceptibility across Caucasians. Though the outcome of the present study was quite in line with the former, there still existed several notable differences. First of all, we attempted to conduct a more comprehensive research to find the underlying association between rs12901499 and OA. Hence, we incorporated several latest published studies with populations beyond Caucasians and conducted subgroup analysis by ethnicity to investigate whether there existed an ethnicity-specific effect of the association. Besides, for each of the included studies, we performed quality assessment with the NOS, which enabled us to judge potential risk of bias as well as grade the level of evidence. After that, considering genetic association studies might be overrepresented by underpowered studies, which indicated a significant association, the power calculation was employed to minimize the possibility of false negatives in the present study, and studies with low power were excluded from the final data synthesis. Last but not least, we carried out sensitivity analysis to validate stability and robustness of the outcome. As a result, all these advantages strongly support us to guarantee a more accurate and reliable conclusion.
Although the present meta-analysis has collected currently available data and provided a more comprehensive evaluation and precise evidence on the association between rs12901499 polymorphism and OA susceptibility, there were several limitations that could not be neglected. First, we only looked into the role of a single loci polymorphism in SMAD3 gene. But as with other complex disorders, OA is modulated by multiple loci polymorphisms and a range of genes with synergetic effects. Therefore, the effects of loci–loci and gene–gene interactions required to be addressed. Second, only 8 articles in English from 4 databanks were retrieved for the meta-analysis and the funnel plot suggested a significant publication bias, potential relevant articles published in other languages and databases might have been missed, which could lead to inclusion criteria bias.38 Third, there was only 1 cohort focused on OA of the hand, spine, as well as TMJ in our study. Therefore, more investigations concerning these sites of OA are necessary to validate the association. Finally, despite the meta-analysis only incorporated case–control designed studies, no study had a satisfying match on age, sex, body mass index, or other confounding factors, which might have caused serious confounding bias in the present study.
Like dozens of other diseases, investigations focused on association of SNPs and OA have been going on for over 2 decades. A growing number of novel loci have been determined, and more are being discovered, which tend to provide new hints and shed light on the underlying mechanisms that affect OA susceptibility. Nevertheless, deviating from original expectation of researchers, most SNPs appear to have only a slight effect on OA, and almost any SNP on hand only seem to increase the risk of OA. In reality, present findings and advances in the genetic basic of OA are nothing more than initial insights into the uncharted territory, and are still far from application in clinical practice of early diagnosis and personalized therapy of OA. As the reach of science always turns ahead in the twists, through unremitting endeavor of researchers, we believe current and future findings will contribute to drive the prevention and treatment of OA through screening individuals if a first-degree relative has a history of OA, or by developing new drugs that target the specific loci.
Conclusion
Collectively, the combined result evidently supported rs12901499 polymorphism in SMAD3 gene was significantly associated with OA vulnerability across both Caucasian and Asian populations. However, the gene polymorphisms identified hitherto were associated with only a fraction of increase in OA risk, and therefore, the determinate etiology of OA may continue to elude researchers. Further work is still encouraged to reveal the etiopathogenesis of OA.
Disclosure
The authors report no conflicts of interest in this work.
References
Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012;64(6):1697–1707. | ||
Nelson AE. Osteoarthritis year in review 2017: clinical. Osteoarthritis Cartilage. 2018;26(3):319–325. | ||
Centers for Disease Control and Prevention (CDC). National and state medical expenditures and lost earnings attributable to arthritis and other rheumatic conditions-United States, 2003. MMWR Morb Mortal Wkly Rep. 2007;56(1):4–7. | ||
Hunter DJ, Schofield D, Callander E. The individual and socioeconomic impact of osteoarthritis. Nat Rev Rheumatol. 2014;10(7):437–441. | ||
Prieto-Alhambra D, Judge A, Javaid MK, Cooper C, Diez-Perez A, Arden NK. Incidence and risk factors for clinically diagnosed knee, hip and hand osteoarthritis: influences of age, gender and osteoarthritis affecting other joints. Ann Rheum Dis. 2014;73(9):1659–1664. | ||
Srikanth VK, Fryer JL, Zhai G, Winzenberg TM, Hosmer D, Jones G. A meta-analysis of sex differences prevalence, incidence and severity of osteoarthritis. Osteoarthritis Cartilage. 2005;13(9):769–781. | ||
Jiang L, Tian W, Wang Y, et al. Body mass index and susceptibility to knee osteoarthritis: a systematic review and meta-analysis. Joint Bone Spine. 2012;79(3):291–297. | ||
Reynard LN, Loughlin J. The genetics and functional analysis of primary osteoarthritis susceptibility. Expert Rev Mol Med. 2013;15:e2. | ||
Hirsch R, Lethbridge-Cejku M, Hanson R, et al. Familial aggregation of osteoarthritis: data from the Baltimore Longitudinal Study on Aging. Arthritis Rheum. 1998;41(7):1227–1232. | ||
MacGregor AJ, Li Q, Spector TD, Williams FM. The genetic influence on radiographic osteoarthritis is site specific at the hand, hip and knee. Rheumatology (Oxford). 2009;48(3):277–280. | ||
Dai X, Wang C, Dai J, et al. Association of single nucleotide polymorphisms in estrogen receptor alpha gene with susceptibility to knee osteoarthritis: a case-control study in a Chinese Han population. Biomed Res Int. 2014;2014:151457. | ||
Lee SW, Song JH, Choi WS, et al. The single nucleotide polymorphism (SNP) of the estrogen receptor-beta gene, rs1256049, is associated with knee osteoarthritis in Korean population. Knee. 2014;21(1):242–246. | ||
Mototani H, Mabuchi A, Saito S, et al. A functional single nucleotide polymorphism in the core promoter region of CALM1 is associated with hip osteoarthritis in Japanese. Hum Mol Genet. 2005;14(8):1009–1017. | ||
Mototani H, Iida A, Nakamura Y, Ikegawa S. Identification of sequence polymorphisms in CALM2 and analysis of association with hip osteoarthritis in a Japanese population. J Bone Miner Metab. 2010;28(5):547–553. | ||
Zhu ZH, Jin XZ, Zhang W, et al. Associations between vitamin D receptor gene polymorphisms and osteoarthritis: an updated meta-analysis. Rheumatology (Oxford). 2014;53(6):998–1008. | ||
Lv ZT, Liang S, Huang XJ, Cheng P, Zhu WT, Chen AM. Association between ADAM12 single-nucleotide polymorphisms and knee osteoarthritis: a meta-analysis. Biomed Res Int. 2017;2017:5398181. | ||
Miyamoto Y, Mabuchi A, Shi D, et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39(4):529–533. | ||
Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-beta/SMAD3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153(1):35–46. | ||
Yao JY, Wang Y, An J, et al. Mutation analysis of the SMAD3 gene in human osteoarthritis. Eur J Hum Genet. 2003;11(9):714–717. | ||
Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. | ||
Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed January 1, 2018. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Jiang LY, Tao YC, Wang YC, et al. A SMAD3 gene polymorphism is related with osteoarthritis in a Northeast Chinese population. Rheumatol Int. 2013;33(7):1763–1768. | ||
Liva E, Panagiotou I, Palikyras S, et al. Candidate gene investigation of spinal degenerative osteoarthritis in Greek population. Spine J. 2017;17(12):1881–1888. | ||
Sharma AC, Srivastava RN, Srivastava SR, Parmar D, Singh A, Raj S. Association between single nucleotide polymorphisms of SMAD3 and BMP5 with the risk of knee osteoarthritis. J Clin Diagn Res. 2017;11(6):GC01–GC04. | ||
Su SL, Yang HY, Lee HS, et al. Gene-gene interactions between TGF-beta/SMAD3 signalling pathway polymorphisms affect susceptibility to knee osteoarthritis. BMJ Open. 2015;5(6):e007931. | ||
Valdes AM, Spector TD, Tamm A, et al. Genetic variation in the SMAD3 gene is associated with hip and knee osteoarthritis. Arthritis Rheum. 2010;62(8):2347–2352. | ||
Xiao JL, Meng JH, Gan YH, Zhou CY, Ma XC. Association of GDF5, SMAD3 and RUNX2 polymorphisms with temporomandibular joint osteoarthritis in female Han Chinese. J Oral Rehabil. 2015;42(7):529–536. | ||
Zhang L, Zhang L, Zhang H, Wang W, Zhao Y. Association between SMAD3 gene rs12901499 polymorphism and knee osteoarthritis in a Chinese population. J Clin Lab Anal. Epub 2018 Jan 8. | ||
Zhong F, Lu J, Wang Y, Song H. Genetic variation of SMAD3 is associated with hip osteoarthritis in a Chinese Han population. J Int Med Res. 2018;46(3):1178–1186. | ||
Heldin CH, Miyazono K, Ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390(6659):465–471. | ||
Roberts AB, Russo A, Felici A, Flanders KC. SMAD3: a key player in pathogenetic mechanisms dependent on TGF-beta. Ann N Y Acad Sci. 2003;995:1–10. | ||
Lee YH, Rho YH, Choi SJ, Ji JD, Song GG. Osteoarthritis susceptibility loci defined by genome scan meta-analysis. Rheumatol Int. 2006;26(11):959–963. | ||
Massague J, Seoane J, Wotton D. SMAD transcription factors. Genes Dev. 2005;19(23):2783–2810. | ||
Li TF, Darowish M, Zuscik MJ, et al. SMAD3-deficient chondrocytes have enhanced BMP signaling and accelerated differentiation. J Bone Miner Res. 2006;21(1):4–16. | ||
van de Laar IM, Oldenburg RA, Pals G, et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet. 2011;43(2):121–126. | ||
Egger M, Smith GD. Bias in location and selection of studies. BMJ. 1998;316(7124):61–66. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




