Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
The Association Between Azathioprine Genetic Polymorphisms, Clinical Efficacy and Adverse Drug Reactions Among Egyptian Patients with Autoimmune Diseases
Authors Abuelsoud N , Fayed H, Elkateeb E
Received 3 October 2020
Accepted for publication 1 December 2020
Published 2 February 2021 Volume 2021:14 Pages 179—187
DOI https://doi.org/10.2147/PGPM.S285033
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Nermeen Abuelsoud,1,2 Hala Fayed,3 Engy Elkateeb4
1Department of Clinical Pharmacy Practice, Faculty of Pharmacy, The British University in Egypt, Cairo, Egypt; 2Department of Pharmacy Practice/Clinical Pharmacy, Faculty of Pharmacy, Egyptian Russian University, Cairo, Egypt; 3Department of Rheumatology and Rehabilitation, Kasr Alaini University Hospital, Faculty of Medicine, Cairo University, Cairo, Egypt; 4Department of Chemical and Clinical Pathology, Kasr Alaini University Hospital, Faculty of Medicine, Cairo University, Cairo, Egypt
Correspondence: Nermeen Abuelsoud Misr-Ismailia Road, PO Box 43, El Sherouk City, 11837, Egypt
Tel +00201226117118
Email [email protected]
Purpose: The study aimed to detect the frequencies of allelic variants (TPMT*3A, TPMT*3C, and TPMT*3G) in the TPMT genes in the Egyptian population and assess the association between TPMT polymorphisms and azathioprine (AZA)—clinical efficacy and adverse drug reactions among Egyptian patients with autoimmune diseases.
Patients and Methods::
Design: A prospective, observational single-center clinical trial.
Setting: Rheumatology and Rehabilitation Department, Kasr Alainy University Hospital, Faculty of Medicine, Cairo University.
Patients: Patients attending Kasr Alainy Rheumatology Outpatient Clinic between December 1, 2017 and June 30, 2019 were included in the study after signing a consent form. TPMT genetic polymorphisms were detected for all patients, and the association between polymorphisms presence and azathioprine’s clinical efficacy and adverse drug reactions were determined.
Results: A total of 150 patients with a mean age of 35.85 years were enrolled in this study. About 72% of patients were heterozygous in the TPMT*3 G460A and TPMT*3 A719G mutant alleles and 81% were wild type in the TPMT*2 G238C mutant allele. Abnormal liver function tests were detected in 42% of patients. Myelosuppression was presented as anemia which was detected in 63% of patients, leucopenia in 51%, and thrombocytopenia in 25% of patients. AZA clinical failure has occurred in 50% of patients where AZA was discontinued or shifted to another drug which occurred in 45% of patients. Myelosuppression rates were higher in homozygous patients in the three mutant alleles, but statistically significant in TPMT*2 G238C while not statistically significant in TPMT*3 G460A and TPMT*3 A719G. Females had a higher risk of immunosuppression than males (p-value 0.031).
Conclusion: The study provided an overview of the genomic variations in the Egyptian population. Routine TPMT genotyping prior to the initiation of AZA therapy should be considered.
Keywords: azathioprine, genetic polymorphisms, autoimmune diseases, Egyptian patients
Introduction
Autoimmune disorders are a group of heterogeneous diseases that occur when the body tissues are attacked by its own immune system.1 During the pathogenesis of these disorders, both T cells and B cells are activated even without any precipitating cause or infection.2 The guidelines for treating these disorders aimed to change the thresholds of immune activation by using immunosuppressants.3 Azathioprine (AZA) is used in the treatment of various autoimmune diseases including systemic lupus erythematosus (SLE). Despite AZA efficacy in treating manifestations of autoimmune diseases, its use is associated with myelosuppression, gastric toxicity, pancreatitis, and liver toxicity. Inter-individual variability to AZA effect and outcomes is related to its genetic polymorphism. AZA is activated by the liver to 6-mercaptopurine (6-MP) through a glutathione-dependent process.4
Thiopurine S-methyltransferase (TPMT) is the enzyme responsible for catalyzing the methylation of 6-MP. It competes with xanthine oxidase and hypoxanthine-guanine phosphor-ribosyl-transferase to detect the percent of 6-MP metabolized to the active metabolite 6-thioguanine nucleotide (6-TGN) metabolite: the main component responsible for immunosuppressive effect.5 Many studies have proved that there are many genetic polymorphisms in the gene encoding for TPMT. Recently, the number of mutations in this specific gene reached 37 mutations.6 Patients were categorized into: (i) intermediate TPMT activity which is heterozygous for a mutant TPMT allele represents 4–11% of patients. (ii) very low or absent TPMT activity which is homozygous or compounds heterozygous about 1 in 300 patients.7–9 Intermediate TPMT activity patients can accumulate 50% more from 6-TGNs in comparison with patients that have normal or high TPMT activity. These individuals will have a high risk of adverse drug reactions (ADRs) caused by AZA. High doses of 6-TGNs will be accumulated in patients with deficient TPMT activity which may result in fatal myelosuppression.10 Another study concluded that very few patients who have two inactive TPMT alleles are homozygous deficient, experienced severe bone marrow suppression; a large percentage of those who are heterozygous encounter moderate to severe bone marrow suppression and those who are homozygous for wild type TPMT alleles have reduced the risk of bone marrow suppression.11 Detecting TPMT genotypes or phenotypes before starting AZA therapy is documented in many updated treatment protocols.11–13 In autoimmune diseases, patients with homozygous wild type should receive full initial doses, while heterozygous patients required lower doses (30–70% of the recommended dose) and substantially very small doses or other drugs are required for the rare homozygous-deficient patients.14 TPMT protective testing has a great advantage as the doses can be tolerated on the basis of TPMT status. This decreases the probability of acute myelosuppression without affecting disease progression.15 To our knowledge, only one study investigated the frequency of TPMT gene polymorphisms (TPMT*3A and TPMT*3C alleles) in Egypt.16 However, no studies investigated the frequency of TPMT*3G or the association between TPMT polymorphisms and AZA clinical efficacy and ADRs among Egyptian patients with autoimmune diseases. This study aimed to detect the frequencies of allelic variants (TPMT*3A, TPMT*3C and TPMT*3G) in the TPMT genes among Egyptian population and to assess the association between TPMT polymorphisms and AZA clinical efficacy and ADRs in Egyptian patients with autoimmune diseases.
Materials and Methods
Design
A prospective, observational single-center clinical trial.
Setting
Rheumatology and Rehabilitation Department, Kasr Alainy University hospital, Faculty of Medicine, Cairo University.
Patients
Patients attending Kasr Alainy Rheumatology Outpatient Clinic between December 1, 2017 and June 30, 2019 was included in the study after signing a consent form. Patients were diagnosed as having an autoimmune/chronic inflammatory disease; systemic lupus erythematosus, Behçet's disease, mixed connective tissue disease, rheumatoid arthritis, and scleroderma during the study period were included in the study after signing a consent form. TPMT genetic polymorphisms were detected for all patients and the association between the presence of polymorphisms, clinical efficacy of AZA and adverse drug reactions were determined.
Exclusion Criteria
Patients were excluded from the study if they had myelosuppression or abnormal liver function tests before receiving AZA.
DNA Isolation and Analysis of TPMT Gene Polymorphisms
A volume of 5 mL of EDTA anticoagulated whole blood was obtained from each patient, genomic DNA was then extracted from peripheral blood nucleated cells. TPMT gene polymorphisms were detected by using real time-polymerase chain reaction (RT-PCR). All samples were analyzed to detect the G238C (rs 1800462) transversion in TPMT*2, the G460A (rs 1800460), and A719G (rs 1142345) transitions in TPMT*3 alleles. Genotypes were determined by using a TaqMan® genotyping assay.
Clinical Data Collection and Patient Follow-up
All patients’ medical files were reviewed to detect the AZA doses and its associated ADRs as well as any change in the treatment plans. Follow-up of all patients continued until the end of the study (April 30, 2020).
Myelosuppression Laboratory Values Used During Analysis
Neutropenia: ANC (absolute neutrophil count) <2000 cells/mm3
Mild neutropenia: ANC between 1000 and 1500 cells/mm3
Moderate neutropenia: ANC between 500and 1000 cells/mm3
Severe neutropenia: ANC <500 cells/mm3
Thrombocytopenia: a platelet count <100000/µL
Mild anemia: hemoglobin between 9.5 and 10.9 g/dL
Moderate anemia: hemoglobin between 8 and 9.4 g/dL
Severe anemia: hemoglobin between 6.5 and 7.9 g/dL
Life-threatening anemia: hemoglobin <6.5 g/dL
Statistical Analysis
Data were analyzed using IBM SPSS Statistics Version 22. Data were tested for normality using the Kolmogorov–Smirnov test and the Shapiro–Wilk test. Quantitative data were presented as mean and standard deviation while qualitative data were presented as number and percentage. Analysis of variance test was used to compare numerical variables between the different categories of SNPs. Chi-squared test or Fisher's exact test as appropriate were used to compare categorical variables between the different categories of SNPs. P-value set significant at 0.05 levels. All tests were two-tailed.
Ethical Approval
The study was approved by the ethical committee for grants of the British University in Egypt.
This study was conducted in accordance with the Declaration of Helsinki.
Results
This study investigated genetic polymorphism in the TPMT gene in a total of 150 patients with a mean age of 35.85 years. Table 1 shows the demographics and patients’ characteristics. The study determined the prevalence of three mutant alleles; TPMT*3 G460A (rs 1800460), TPMT*2 G238C (rs 1800462), and TPMT*3 A719G (rs 1142345). About 72% of patients were heterozygous in the TPMT*3 G460A and TPMT*3 A719G mutant alleles and 81% were wild type in the TPMT*2 G238C mutant allele as shown in Table 1. Seventy percent of patients’ diagnoses were systemic lupus erythematosus. A dose of 100 mg (1*2=one 50 mg tablet *2) was the initial dose in 94% of patients and about 75% of patients were maintained on the same dose. Abnormal liver function tests were detected in 42% of patients. Myelosuppression was represented as anemia which was detected in 63% of patients, leucopenia in 51%, and thrombocytopenia in 25% of patients. The AZA-clinical failure occurred in 50% of patients where AZA discontinuation or shift to another drug (in 45% of patients).
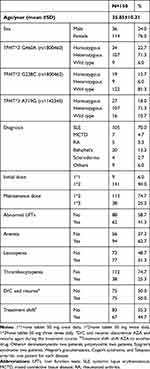 |
Table 1 Demographics and Patients Characteristics |
Table 2 shows the frequencies of homozygous, heterozygous, and wild type carriers of a mutant allele of the TPMT*3 G460A. Myelosuppression was presented as anemia and leucopenia and there was a statistically significant difference between the homozygous, heterozygous and wild type carriers of the mutant allele of the TPMT*3 G460A as shown in Table 2. There was also a statistically significant difference between the three types in regard to the initial and maintenance doses, the severity of anemia as well as the treatment shift parameter.
 |
Table 2 AZA Clinical Efficacy and Associated ADRs in Different Types of TPMT *3 G460A Mutant Allele |
Table 3 shows the frequencies of homozygous, heterozygous, and wild type carriers of the mutant allele of the TPMT*2 G238C. Myelosuppression was presented as anemia and leucopenia and there was a statistically significant difference between the homozygous, heterozygous, and wild-type carriers of the mutant allele of the TPMT*2 G238C as shown in Table 3. There was also a statistically significant difference between the three types in regard to the severity of anemia as well as the treatment shift parameter.
 |
Table 3 AZA Clinical Efficacy and Associated ADRs in Different Types of TPMT 2* G238C Mutant Allele |
Table 4 shows the frequencies of homozygous, heterozygous, and wildtype carriers of the mutant allele of the TPMT*3 A719G. Abnormal liver function tests (LFTs) were presented as elevation in the aspartate transaminase (AST) and alanine transaminase (ALT) and there was a statistically significant difference between the homozygous, heterozygous and wild type carriers of the mutant allele of the TPMT*3 A719G as shown in Table 4. There was also a statistically significant difference between the three types in regard to the treatment shift parameter. Myelosuppression rates were higher in homozygous patients in the three mutant alleles, but it was statistically significant in TPMT*2 G238C while not statistically significant in TPMT*3 G460A and TPMT*3 A719G. Females had a higher risk of immunosuppression than males (p-value 0.031) as shown in Table 5.
 |
Table 4 AZA Clinical Efficacy and Associated ADRs in Different Types of TPMT 3* A719G Mutant Allele |
 |
Table 5 Myelosuppression Rates in the Different Types of TPMT Mutant Alleles |
Discussion
This current study detected the frequencies of three mutant TPMT alleles; TPMT*3 G460A, TPMT*2 G238C, and TPMT*3 A719G. About 72% of patients were heterozygous in the TPMT*3 G460A and TPMT*3 A719G mutant alleles and 81% were wildtype in the TPMT*2 G238C mutant allele. These findings are consistent with the results of many other studies conducted by Kubota and Chiba; Kumagai et al; and Hiratsuka et al, these studies showed that in Caucasians TPMT*3A (460G>A: rs1800460 and 719A>G: rs1142345) represents the most frequent allele, whereas TPMT*3C (719A>G: rs1142345) represents the most frequent allele in the Japanese population.17–19 In this current study, TPMT*3 G460A, TPMT*2 G238C, and TPMT*3 A719G are the most common defective alleles of the TPMT gene examined. About 23% of patients were homozygous in the TPMT*3 G460A mutant allele; the rates of heterozygous and wild type were 71% and 6%, respectively. The percentages of homozygous, heterozygous and wild type in the mutant allele TPMT*2 G238C was 13, 6, and 81%, respectively, while these rates were 18, 71, and 11% in the TPMT*3 A719G mutant allele.
Our results were discordant with a study conducted on 194 Egyptian patients done by Hamdy et al that concluded that the frequency of wild type allele was 97% and 3% of patients who were heterozygous for one of the mutant alleles. TPMT*1/TPMT*3A was detected in one patient, while TPMT*1/TPMT*3C was detected in five subjects. Neither G238C (TPMT*2), G460A alone (TPMT*3B) nor the homozygous type of any mutant allele was determined in the Egyptian population. Their study also concluded that TPMT*3C is the inherited TPMT variant allele in the Egyptian population, accounting for 86% of the variant alleles detected. TPMT*3A is the second most recurrent variant and accounts for the remaining 14% of the TPMT variant alleles in Egyptians.16
In comparison with other global populations across Asia, Europe and the Americas, the gene variant that occurs most frequently in Caucasians is TPMT*3A (approximately 85%), *2, *3B and *3C occur much less frequently. In Africans and African Americans, as in Caucasians, *1 is the wild type, but the most common allele is TPMT*3C.20,21 Regarding the Asian population, TPMT*3C is also the most common mutation reported while, TPMT*3A does not occur.22–24 TPMT activity, in addition to nudix hydrolase 15 genetics, is a critical factor contributing to thiopurine-induced hematotoxicity.25 TPMT activity differs among patients as about 87% of patients have high TPMT activity, 11.1% have intermediate activity, and 0.3% have low or absent activity.26–28 Patients with normal TPMT activity are categorized as the wild type TPMT*1.29 Patients (~1 in 178 to 1 in 3736) who are homozygous-deficient universally develop severe bone marrow suppression; an increased percentage of those who are heterozygous develop moderate-to-severe bone marrow suppression, and those who are homozygous for wild type TPMT alleles are at reduced risk of myelosuppression.11 These results were consistent with our findings as 72.5% of homozygous-type carriers of the mutant allele TPMT*3 G460A developed anemia and leucopenia. Anemia and leucopenia represented 80% and 84% of homozygous-type carriers of the mutant allele TPMT*2 G238C and 59% for each of them of homozygous-type carriers of the mutant allele TPMT*3 A719G.
Myelosuppression was presented as anemia, leucopenia, or thrombocytopenia; these rates were higher in homozygous patients in the three mutant alleles, but it was statistically significant in TPMT*2 G238C while it was not statistically significant in TPMT*3 G460A and TPMT*3 A719G. This study showed ADR as well as another study that concluded that patients who inherit two nonfunctional TPMT alleles are at 100% risk for life-threatening bone marrow suppression and that heterozygous patients cannot tolerate full doses of AZA.30,31 This current study showed that females had a higher risk of immunosuppression than males (p-value 0.031). This finding is supported by many other studies that concluded that TPMT activity increases in children compared with adults, between adults, it is elevated in males rather than in females.32–34 During patients follow-up, AZA was discontinued in 50% of patients due to ADRs development, while treatment shift occurred in 45% of patients. Another retrospective study was conducted in the UK and included 113 patients with inflammatory bowel disease mentioned that AZA discontinuation has resulted from AZA adverse effects.35 In this current study, about 94% of patients initiated on a total of 100 mg dose (<2 mg/kg) and 75% of patients could not tolerate a maintenance dose more than this dosing range. Our findings were supported by another study which concluded that there was a good clinical response in patients who received low doses of azathioprine (<2 mg/kg). Relapse or lack of response was associated with using of low-AZA dose in patients with high TPMT activity.36
Conclusion
The study provided an overview of the genomic variations in the Egyptian population. The therapy should be started with a low dose of azathioprine, slowly increase the dose—under close blood counts—until an optimal effect is achieved without the occurrence of side effects.
Routine TPMT genotyping prior to the initiation of AZA therapy may be helpful for Egyptian patients in order to determine whether the drug is appropriate for each patient. Further studies should be conducted to compare the cost-effectiveness of prior screening before starting treatment and treating patients without screening in the Egyptian population. Genotyping will likely provide great pharmacologic and economic benefits in countries like Egypt.
Acknowledgments
This work was supported by the British University in Egypt.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Das PK, Elliott G. Conference scene: lessons from animal models of autoimmune diseases: from mechanisms to applications. Immunotherapy. 2011;3(2):147–151. doi:10.2217/imt.10.102
2. Sinha AA, Lopez MT, McDevitt HO. Autoimmune diseases: the failure of self tolerance. Science. 1990;248(4961):1380–1388. doi:10.1126/science.1972595
3. Goding JW. Autoimmune diseases. N Engl J Med. 2001;345(23):1707–1708.
4. Lennard L. The clinical pharmacology of 6-mercaptopurine. Eur J Clin Pharmacol. 1992;43(4):329–339. doi:10.1007/BF02220605
5. Lennard L. TPMT in the treatment of Crohn’s disease with azathioprine. Gut. 2002;51(2):143–146. doi:10.1136/gut.51.2.143
6. Appell ML, Berg J, Duley J, et al. Nomenclature for alleles of the thiopurine methyltransferase gene. Pharmacogenet Genomics. 2013;23(4):242–248. doi:10.1097/FPC.0b013e32835f1cc0
7. Collie-Duguid ES, Pritchard P, Powrie P, et al. The frequency and distribution of thiopurine methyltransferase alleles in Caucasian and Asian populations. Pharmacogenetics. 1999;9(1):37–42.
8. Engen RM, Marsh S, Van Booven DJ, et al. Ethnic differences in pharmacogenetically relevant genes. Curr Drug Targets. 2006;7(12):1641–1648. doi:10.2174/138945006779025446
9. Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity.. Am J Hum Genet. 1980;32(5):651–662.
10. Egan LJ, Derijks LJJ, Hommes DW. Pharmacogenomics in inflammatory bowel disease. Clin Gastroenterol Hepatol. 2006;4(1):21–28. doi:10.1016/j.cgh.2005.10.003
11. Relling MV, Gardner EE, Sandborn WJ, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther. 2011;89(3):387–391. doi:10.1038/clpt.2010.320
12. Relling MV, Gardner EE, Sandborn WJ, et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther. 2013;93(4):324–325. doi:10.1038/clpt.2013.4
13. Anstey AV, Wakelin S, Reynolds NJ. British Association of Dermatologists Therapy G, Audit S. Guidelines for prescribing azathioprine in dermatology. Br J Dermatol. 2004;151(6):1123–1132. doi:10.1111/j.1365-2133.2004.06323.x
14. Ford LT, Berg JD. Thiopurine S-methyltransferase (TPMT) assessment prior to starting thiopurine drug treatment; a pharmacogenomic test whose time has come. J Clin Pathol. 2010;63:288–295. doi:10.1136/jcp.2009.069252
15. Schmiegelow K, Forestier E, Hellebostad M, et al. Long-term results of NOPHO ALL-92 and ALL-2000 studies of childhood acute lymphoblastic leukemia. Leukemia. 2010;24(2):345–354. doi:10.1038/leu.2009.251
16. Hamdy SI, Hiratsuka M, Narahara K, et al. Genotype and allele frequencies of TPMT, GST, SULT1A1 and MDR-1in the Egyptian population. British Journal of Clinical Pharmacology. 2003;55(6):560–569. doi:10.1046/j.1365-2125.2003.01786.x
17. Kubota T, Chiba K. Frequencies of thiopurine S-methyltransferase mutant alleles (TPMT*2, *3A, *3B and *3C) in 151 healthy Japanese subjects and the inheritance of TPMT*3C in the family of a propositus. Br J Clin Pharmacol. 2001;51(5):475–477.
18. Kumagai K, Hiyama K, Ishioka S, et al. Allelotype frequency of the thiopurine methyltransferase (TPMT) gene in Japanese. Pharmacogenetics. 2001;11(3):275–278. doi:10.1097/00008571-200104000-00012
19. Hiratsuka M, Inoue T, Omori F, et al. Genetic analysis of thiopurine methyltransferase polymorphism in a Japanese population. Mutat Res. 2000;448(1):91–95. doi:10.1016/S0027-5107(00)00004-X
20. Ameyaw MM. Thiopurine methyltransferase alleles in British and Ghanaian populations. Hum Mol Genet. 1999;8(2):367–370. doi:10.1093/hmg/8.2.367
21. Hon YY, et al. Polymorphism of the thiopurine S-methyltransferase gene in African- Americans. Hum Mol Genet. 1999;8(2):371–376. doi:10.1093/hmg/8.2.371
22. Chang J, Lee L, Chen C, et al. Molecular analysis of thiopurine S-methyltransferase alleles in South-east Asian populations. Pharmacogenetics. 2002;12(3):191–195. doi:10.1097/00008571-200204000-00003
23. Collie-Duguid ESR, Pritchard P. The frequency and distribution of thiopurine methyltransferase alleles in Caucasian and Asian populations. Pharmacogenetics. 1999;9(1):37–42. doi:10.1097/00008571-199902000-00006
24. Kubota T, Chiba K. Frequencies of thiopurine S-methyltransferase mutant alleles (TPMT*2, *3A, *3B and *3C) in 151 healthy Japanese subjects and the inheritance of TPMT*3C in the family of a propositus. British Journal of Clinical Pharmacology. 2001;51(5):475–477. doi:10.1046/j.1365-2125.2001.01371.x
25. Schaeffeler E, Jaeger SU, Klumpp V, et al. Impact of NUDT15 genetics on severe thiopurine-related hematotoxicity in patients with European ancestry. Genet Med. 2019;21(9):2145–2150. doi:10.1038/s41436-019-0448-7
26. Siva C. Pharmacogenetics in rheumatology: the prospects and limitations of an emerging field. Rheumatology. 2002;41(11):1273–1279. doi:10.1093/rheumatology/41.11.1273
27. Weinshilboum R, Guttmacher AE, Collins FS. Inheritance and drug response. N Engl J Med. 2003;348(6):529–537. doi:10.1056/NEJMra020021
28. Arnott IDR, Watts D, Satsangi J. Azathioprine and anti-TNFα therapies in Crohn’s disease: a review of pharmacology, clinical efficacy and safety. Pharmacol Res. 2003;47(1):1–10. doi:10.1016/S1043-6618(02)00264-5
29. McLeod HL, Siva C. The thiopurine S-methyltransferase gene locus – implications for clinical pharmacogenomics. Pharmacogenomics. 2002;3(1):89–98. doi:10.1517/14622416.3.1.89
30. Evans WE, Hon YY, Bomgaars L, et al. Preponderance of thiopurine S-methyltransferase deficiency and heterozygosity among patients intolerant to mercaptopurine or azathioprine. Journal of Clinical Oncology. 2001;19(8):2293–2301. doi:10.1200/JCO.2001.19.8.2293
31. Stocco G, Cheok MH, Crews KR, et al. Genetic polymorphism of inosine triphosphate pyrophosphatase is a determinant of mercaptopurine metabolism and toxicity during treatment for acute lymphoblastic leukemia. Clinical Pharmacology & Therapeutics. 2009;85(2):164–172. doi:10.1038/clpt.2008.154
32. Schaeffeler E, Fischer C, Brockmeier D, et al. Comprehensive analysis of thiopurine S-methyltransferase phenotype???genotype correlation in a large population of German-Caucasians and identification of novel TPMT variants. Pharmacogenetics. 2004;14(7):407–417. doi:10.1097/01.fpc.0000114745.08559.db
33. Pettersson B, Almer S, Albertioni F, et al. Differences between children and adults in thiopurine methyltransferase activity and metabolite formation during thiopurine therapy: possible role of concomitant methotrexate. Ther Drug Monit. 2002;24(3):351–358. doi:10.1097/00007691-200206000-00005
34. Indjova D, Atanasova S, Shipkova M, et al. Phenotypic and genotypic analysis of thiopurine s-methyltransferase polymorphism in the bulgarian population. Ther Drug Monit. 2003;25(5):631–636. doi:10.1097/00007691-200310000-00013
35. Baker DE. Pharmacogenomics of azathioprine and 6-mercaptopurine in gastroenterologic therapy.. Rev Gastroenterol Disord. 2003;3(3):150–157.
36. Campbell S, Kingstone K, Ghosh S. Relevance of thiopurine methyltransferase activity in inflammatory bowel disease patients maintained on low-dose azathioprine. Aliment Pharmacol Ther. 2002;16(3):389–398. doi:10.1046/j.1365-2036.2002.01177.x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
