Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 13
TGF-β1 Gene Polymorphism and Its Correlation with Serum Level of TGF-β1 in Psoriasis Vulgaris Among Iraqi People
Authors Ahmed BT , Saeed MY, Noori SH, Amin DM
Received 16 September 2020
Accepted for publication 18 November 2020
Published 24 November 2020 Volume 2020:13 Pages 889—896
DOI https://doi.org/10.2147/CCID.S281585
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Bryar T Ahmed,1 Mohammad Y Saeed,1 Saman H Noori,2 Dashty M Amin3
1Department of Medicine/Dermatology, College of Medicine, University of Sulaimani, Sulaimani City, Kurdistan, Iraq; 2Department of Biochemistry, College of Medicine, University of Sulaimani, Sulaimani City, Kurdistan, Iraq; 3Medical Laboratory Sciences, Komar University of Science and Technology, Sulaimani City, Kurdistan, Iraq
Correspondence: Bryar T Ahmed
Department of Medicine/Dermatology, College of Medicine, University of Sulaimani, P.O Box 97, Hawarabarza, Sulaimani City, Iraq
Tel +9647701520468
Email [email protected]
Purpose: Many cytokines have been implicated in the pathogenesis of psoriasis, among these the transforming growth factor-beta 1 (TGF-β 1) can be endorsed by different mechanisms besides inhibiting keratinocytes proliferation. The role of genetic polymorphisms of TGF-β 1 has been studied in various inflammatory diseases. Our aim is to study the correlation of TGF-β 1 gene polymorphism at codon 10 and 25 with the expression of serum level of TGF-β 1 in a sample of Iraqi psoriatic patients compared to the control group.
Materials and Methods: A cross-sectional study involved 100 patients with psoriasis vulgaris and 50 sex- and age-matched healthy volunteers as control group. Serum and genomic DNA were prepared from peripheral blood samples. Amplification refractory mutation system–polymerase chain reaction technique (ARMS-PCR) had been applied for genotyping TGF-β 1 codon 10 [rs1982073] and codon 25 [rs1800471] genetic polymorphisms. Enzyme-linked immunosorbent assay technique (ELISA) based on the sandwich principle was used for quantification of serum TGF-β 1 level. Psoriasis Area and Severity Index (PASI) scoring was applied for determining the severity in psoriatic patients and classified accordingly to mild (PASI< 7), moderate (PASI 7– 12), severe (PASI> 12) groups.
Results: Statistically significant difference was found in TGF-β 1 gene polymorphism between psoriatic patients and control group at codon 10 (T869C) polymorphism (p=0.021) and codon 25 (G915C) polymorphism (p=0.040). No significant association was detected with the mean serum TGF-β 1 level, severity of the disease, disease onset, gender, history of psoriatic arthritis, and smoking in both codons. Significant lower mean serum TGF-β 1 level was found among psoriatic group (192.17 ± 531.12 ng/L) compared with controls (565.89 ± 1372.30 ng/L) (p = 0.018). Relation of mean serum TGF-β 1 level with the onset of the disease was statistically significant (p = 0.004), early-onset disease group was lower (105.92 ± 68.02 ng/L) compared with the late-onset disease group (450.92 ± 1027.79 ng/L). The mean serum TGF-β 1 level showed no significant differences with the severity of psoriasis, gender, history of psoriatic arthritis, and smoking.
Conclusion: Iraqi population showed a significant association between TGF-β 1 gene polymorphism at codon 10 and 25 were with psoriasis susceptibility, and a significantly lower mean serum TGF-β 1 level was detected in psoriatic patients.
Keywords: psoriasis, TGF-β 1, gene, polymorphism
Introduction
Psoriasis, a chronic inflammatory immune-mediated hyperproliferative disease that affects the skin, has an estimated global prevalence of 2–4%.1,2 The severity varies from a few numbers of distributed red, scaly plaques to a widely scattered that involves most of the body, it may get worse with age, or ultimately increase and decrease in its severity, which depends on inheritance and environmental factors.3 Psoriasis can be classified as early- and late-onset, determined as type I and II, respectively; type I is that begins before age of 40 years, and type II is that begins on or after 40 years of age.4
Keratinocyte hyperplasia in psoriatic lesions may be attributable to change in the regulation of the responsible growth factors of epidermal proliferation and alteration in the metabolism of their receptors.5 Transforming growth factor-beta (TGF-β) is regarded as a dimeric polypeptide growth factor with three isoforms (TGF-β1, TGF-β2, and TGF-β3) that had been determined in human tissues.6 TGF-β1 and TGF-β2 were found in the human epidermis and also present in the dermis, whereas TGF-β3 is found in the dermis, mainly in the upper dermis. It has an inhibitory role on cell proliferation as it stops the cell cycle in the G1 phase and directly enhances angiogenesis. It has been suspected that reduced TGF-β1 signaling potentiates keratinocyte proliferation in psoriatic epidermis.7,8 In the meantime, the increase of TGF-β1 can trigger angiogenesis and stimulates fibroblast proliferation and production of extracellular matrix elements. Moreover, TGF-β1 may increase the production of T-helper1 type cytokine as a type 1 immune response and also has a significant role in the maturation of skin-homing dendritic cells, which is crucial for triggering inflammatory and immune response upon various stimuli.9,10
The human TGF-β1 gene is located on the long arm of chromosome 19; there are five described polymorphisms in this gene: two in the promoter region at positions 800 G/A and 509 C/T and three located in the coding sequence at positions 869 T/C, 915 G/C, and 1628 C/A. The mutation point in position 869 at codon 10 involves alleles T (leucine) and C (proline) and the second one in position 915 at codon 25 G (arginine) and C (proline).11
It has been proved that the ability of a human being to produce a higher or lower level of TGF-β1 may be genetically affected and polymorphisms in its gene regulate TGF-β1 expression.12 These polymorphisms may play in predisposing a person to various disease states, including Rheumatoid arthritis, colorectal carcinoma, diabetes mellitus, osteoporosis, asthma, Crohn’s disease, and fibrotic diseases of the skin and kidney.11,13–17 There are some contradictory results regarding TGF-β1 gene polymorphisms in psoriasis, El-Hadidi et al showed a significant relation between psoriatic patients compared with normal persons as regards codon 10 TGF-β1 gene polymorphism, they concluded that the presence of TT genotype was associated with a threefold risk of psoriasis compared to CC genotype, while another study was done by Baran et al on codon 10 and 25 TGF-β1 gene polymorphism showed no significant relation regarding increasing the liability to psoriasis vulgaris.18,19
Our study aimed to investigate the association between two common polymorphisms in the gene encoding TGF-β1 which were codon 10 in position −869 (T/C) and codon 25 in position −915 (G/C) whether those polymorphisms affect the susceptibility to psoriasis vulgaris and estimating the serum level of TGF-β1 in patients with psoriasis vulgaris to show possible relations with different genotypes in the former polymorphisms, disease severity, age of onset and family history of the same condition.
Materials and Methods
In this cross-sectional study, a hundred patients affected with psoriasis vulgaris were included as the cases group and fifty healthy volunteers served as control group, the whole sample ages ranging from 6 to 77 years old. All patients and controls were recruited from Sulaimani teaching dermatology center and dermatology outpatient clinic in Shorsh general teaching hospital. The study was approved by the medical researches scientific and ethical committee (RSEC-Medical) of the faculty of medical sciences, University of Sulaimani by ethical code (9:28-5-2019), and the study was in accordance with the declaration of Helsinki. Written informed consent was signed by patients and controls whose age was above 18 years and parents had signed the same informed consent for those who were less than 18 years. Inclusion criteria were patients with psoriasis vulgaris of both genders have not received topical or systemic treatment for at least 2 months before collecting a blood sample from them, while exclusion criteria were psoriatic patients rather than psoriasis vulgaris. As regards controls, those with a positive family history of psoriasis, rheumatoid arthritis, colorectal carcinoma, diabetes mellitus, osteoporosis, asthma, Crohn’s disease, fibrotic diseases of the skin and kidney or any other autoimmune diseases have excluded that may affect the mean serum level of TGF-β1. Proper history from patients and controls had been taken, and careful clinical examination of the patients had been done regarding distribution, anatomical sites, and extent of the lesions were evaluated by calculating the Psoriasis Area and Severity Index score (PASI).20 The severity of the disease was classified accordingly to mild (PASI<7), moderate (PASI 7–12), severe (PASI>12).
Eight milliliters of venous blood was withdrawn under complete aseptic conditions from every patient and control. Three milliliters were then collected in sterile ethylene diamine tetra-acetic acid (EDTA) vacationer tubes, and the remaining 5 mL were collected to vacuum blood collection tubes with gel and clot activator to obtain serum, the serum then collected inside a 3-mL Eppendorf tube. The samples, both blood and serum, were labeled and then stored at −20° C.
Detection of TGF-β1 Codon 10, 25 Polymorphism Gene Polymorphism
A genomic deoxyribonucleic acid (DNA) extraction kit (RIBO-prep; AmpliSens, Moscow 111123 Russia) was used to extract DNA from fresh whole blood samples. TGF-β1 codon 10 and 25 genetic polymorphisms were genotyped by using amplification refractory mutation system–polymerase chain reaction (ARMS-PCR) that had been applied for genotyping TGF-β1 codon 10 and 25 genetic polymorphisms in other studies.14,21 ARMS-PCR is a variant of PCR based on the principle that under optimized conditions, the primers with 3′ end mismatch with the complimentary template DNA will not result in the amplification of targeted DNA fragment. For each DNA template, two complementary reactions were established, two sets of primers were used, two forward primers to screen for each allele separately and a common reverse primer which has been supplied by ADD Bio Inc., South Korea.
Primer sequences for codon-10 (T869C) were 5`-GCAGCGGTAGCAGCAGCG-3` specific for C allele, 5`-AGCAGCGGTAGCAGCAGCA-3` specific for T allele, and 5` TCCGTGGGATACTGAGACAC-3` as a generic primer; while for codon-25 (G915C) were 5`-GTGCTGACGCCTGGCCC-3` specific for C allele, 5`-GTGCTGACGCCTGGCCG-3` specific for G allele and 5`-GGCTCCGGTTCTGCACTC-3` as a generic primer. Internal control primers of human growth hormone were used to check that a successful PCR had been run with primer sequences 5`-GCCTTCCCAACCATTCCCTTA-3`, 5`-TCACGGATTTCTGTTGTGTTTC-3` for the forward and reverse primers, respectively.
The PCR reaction was carried out in 20 µL (10 µL master mix from ADD Bio, South Korea, 1 µL of 10 pmol of each forward and reverse primers, 100ng template DNA, and the volume was finished to 10 µL with diethyl-pyrocarbonate treated double-distilled water). PCR was performed using a three-step cycling protocol with initial denaturation for polymerase activation (95ºC/5 min) one cycle; followed by denaturation (95ºC/30 sec), annealing (60ºC/30 sec), extension (72ºC/30 sec) of 40 cycles, and final extension (72ºC/5 sec) of 1 cycle using a thermal cycler (Bio-Rad C1000, USA). A known positive and negative control was included in the run with each batch of amplifications. The amplified PCR product sizes were analyzed by 0.8% agarose gel. PCR 241, 233, and 429 bp for codons 10, 25, and internal control, respectively. The size of target DNA bands compared with 100 bp DNA ladders (GENETBIO) and the gels were documented under ultraviolet light (Ultraviolet Transilluminator, BioDoc-It). dx.doi.org/10.17504/protocols.io.bma5k2g6
Quantification of TGF-β1 in Serum
Enzyme-Linked Immunosorbent Assay technique (ELISA) kit from BT Lab Bio-assay (Shanghai, China), which is based on the sandwich principle, used for accurate quantitative detection of TGF-β1in the serum of patients and controls. The reagent was prepared by reconstituting the 120μL of the standard (4800ng/L) with 120μL of standard diluent to generate a 2400ng/L standard stock solution, then serially diluting the standard stock solution in a 1:2 ratio producing 1200ng/L, 600ng/L, 300ng/L, and 150ng/L solutions. Standard diluent serves as the zero standards (0 ng/L). The washing buffer was prepared by diluting 20mL of wash buffer concentrate 25x into deionized distilled water to yield 500 mL of 1x wash buffer. The procedure was done by adding 50μL standard to standard well, 40μL serum sample and then adding 10μL anti-TGF-β1 antibody to sample wells, then adding 50μL streptavidin-conjugated horseradish peroxidase enzyme to sample wells and standard, incubating for 60 minutes at 37°C. Later on, successive washing the plate 5 times with wash buffer, then adding 50μL substrate solution A, 50μL substrate solution B to each well respectively and incubating the plate for 10 minutes at 37°C in the dark, then finalizing the procedure by adding 50μL stop solution to each well. Determining the optical density (OD value) of each well immediately using a microplate reader set to 450 nm within 10 minutes after adding the stop solution. Plotting the mean absorbance obtained from each standard against its concentration was used to construct the standard curve, which was used to determine the concentration of each sample. The minimum detectable concentration of TGF-β1 by this assay was estimated to be 10ng/L. dx.doi.org/10.17504/protocols.io.bma6k2he
Statistical Analysis
Data were analyzed using the Statistical Package for Social Sciences (SPSS, version 25). The Chi-square test of association was used to compare proportions. Fisher’s exact test was used when the expected count of more than 20% of the cells of the table was less than 5. The Student’s t-test of two independent samples was used to compare two means. One-way analysis of variance22 was used to compare three means. A p-value of ≤ 0.05 was considered statistically significant. Hardy–Weinberg equilibrium (HWE) was tested for patients, controls, and combined groups using the Chi-square test. P-values <0.05 were determined as significant.
Results
A hundred patients with psoriasis vulgaris were included in the study, 48 were males (48%) and 52 were females (52%), the mean age was 42.4 ± 14.8 years, the control group were included 50 healthy volunteers, 31 were males (62%) and 19 were females (38%), the mean age was 46.3 ±19.5 years. The mean age of the whole sample was 43.7 ± 16.6 years, ranging from (6–77) years. The median was 43 years. The demographics of both patients and the control group shown in Table 1. Among the psoriatic group, the early-onset group (onset before the age 40) was 75cases (75%), while 25 cases (25%) had included in the late-onset group (onset on or after the age 40). The duration of the psoriasis was ranged from (0.25–50) years with a median of (10) years. Regarding the degree of severity, 53 (53%) were mild, 19 (19%) were moderate and 28 (28%) were severe according to the PASI scoring. Family history of psoriasis was statistically significant (P<0.001) among psoriatic patients.
 |
Table 1 Demographic Data of Studied Groups |
Studying genotype distribution of TGF-β1 gene polymorphisms among both groups at codon 10 showed a higher proportion of both genotypes; heterozygous T/C (Leucine/Proline; 52%) and homozygous T/T (Leucine/Leuucine; 24%) in the psoriatic group, while a higher proportion of homozygous genotype C/C (Proline/Proline; 46%) was found among controls group, these relations were statistically significant (p=0.021). Nearly the same pattern had been observed in codon 25, which showed a higher proportion of both genotypes; heterozygous G/C (Arginine/Proline; 50%) and homozygous G/G (Arginine/Arginine; 29%) in the psoriatic group, while among the control group higher proportion of homozygous genotype C/C (Proline/Proline; 40%) was found, these relations were also statistically significant (p=0.040) as showed in detail in Table 2. No significant association was detected with the severity of the disease, disease onset, gender, history of psoriatic arthritis, and smoking in both codons.
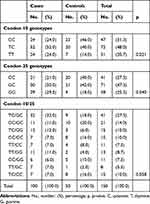 |
Table 2 Genotype Distribution of TGF-β1 Gene Polymorphisms at Codon 10 and 25 of Psoriatic Patients and Controls |
Studying combined codon 10/25 polymorphisms of TGF-β1 gene among both psoriatic and patients group resulted in nine different genotypes as shown in Table 2, the dominant-combined genotypes in the psoriatic group were TC/GC, TC/GG, TT/GG, and TT/GC with the percentage of (32%, 12%, 11%, and 7%) respectively, while the dominant genotypes in control group were CC/GC which were 20%; statistically, the p-value was (0.058).
Genotype frequencies were consistent with HWE in codon 10 (Χ2=0.16, P=0.689 for patients), (Χ2=0.591, P=0.441 for controls) and (Χ2=0.420, P=0.516 for combined group), in codon 25 (Χ2=0.004, P=0.948 for patients), (Χ2=0.687, P=0.406 for controls) and (Χ2=0.420, P=0.516 for combined group).
It is evident in Table 3 that the mean serum TGF-β1 level among the psoriatic group was 192.17 ± 531.12 ng/L, it was significantly (p = 0.018) lower than the mean among the controls 565.89 ± 1372.30 ng/L. The mean serum TGF-β1level was also low among those with the early-onset disease group 105.92 ± 68.02 ng/L compared with the late-onset disease group 450.92 ±1027.79 ng/L and the difference was statistically significant (p = 0.004) as showed in Table 4. No significant differences were detected in the mean serum TGF-β1 level among the different categories of the following variables: severity of psoriasis, gender, history of psoriatic arthritis, and smoking as showed in detail in Table 5.
 |
Table 3 Mean Serum TGF-β1 Level in Psoriatic Patients and Controls |
 |
Table 4 Mean Serum TGF-β1 Level by Psoriasis Age of Onset Categories |
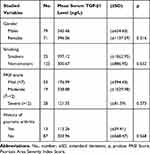 |
Table 5 Mean Serum TGF-β1 Level by Studied Variables |
Studying the relation between means serum TGF-β1 level with genotype distributions of TGF-β1 gene polymorphisms at codon 10 and 25 among both psoriatic and control groups showed no significant difference as showed in Table 6. In codon 10, p-values were 0.538 and 0.707 for psoriatic and control groups, respectively, in codon 25 were 0.650 and 0.125 for the former two groups. Also, the relations of codon 10/25 genotypes with mean serum TGF-β1 level were not significant statistically among both the psoriatic group (p =0.963) and controls (p = 0.566).
 |
Table 6 Relation of Means Serum TGF-β-1 Level with Codon 10, 25 and 10/25 Genotypes |
Discussion
The potential expression of most cytokines could be affected by polymorphic gene sequences of their gene loci and this may have a role in certain inflammatory diseases. TGF-β1 may be genetically affected and polymorphisms in its gene regulate TGF-β1 expression.12 These polymorphisms have been implicated in increase susceptibility of various disease states, including rheumatoid arthritis, colorectal carcinoma, diabetes mellitus, osteoporosis, asthma, fibrotic diseases of the skin and kidney, Crohn’s disease, and systemic lupus erythematosus.11,13–17,23 In this study, a significant difference in TGF-β1 gene polymorphisms was found at codon 10 and 25 between psoriatic patients and controls, both codons showed significant lower frequencies of homozygous (CC) genotypes compared with homozygous (TT), heterozygous (TC) at codon 10 and homozygous (GG), heterozygous (GC) at codon 25, but there was no significant difference with severity and onset of the disease. The same result was found in an Egyptian study regarding codon 10 gene polymorphism in psoriatic patients, which showed a significantly lower proportion of CC genotype.18 In contrast to our finding, a Polish study did not find a difference regarding susceptibility to psoriasis vulgaris and TGF-β1 gene polymorphism in codon 10 and 25, but there was a significantly higher frequency of CC genotype in the early-onset group.19
A significantly lower mean serum level of TGF-β1 was observed in psoriatic patients when compared with the controls in this study, this may be explained by the growth inhibitory effect on keratinocytes that reduced TGF-β1 signaling potentiates keratinocyte hyperproliferation in psoriatic epidermis.24 On the other hand, a study on TGF-β1–deficient mice die from cardiac, pulmonary, and gastric inflammation, suggesting that TGF-β1 has a vital role in suppressing the activation proliferation of inflammatory cells.25 TGF-β1 is a growth factor necessary for the differentiation of Treg cells and, consequently, for the regulation of the immune response.26 Agreements to our results were found in other studies that showed lower serum TGF-β1 in psoriatic patients.27,28 There was no significant difference in our study regarding the relation of mean serum level of TGF-β1 with the disease severity. In contrast to other studies, no significant difference was shown between psoriatic patients compared with the controls, but there was a positive significant relationship with the severity of disease.7,29–31
The relation of mean serum level of TGF-β1 with codon 10 and 25 genotypes did not show any significant difference in this study; the explanation may be that the etiopathogenesis of psoriasis is multifactorial. Unfortunately to the best of our knowledge, no similar studies had been done in this regard. Furthermore, studies on larger samples and in different ethnicities in the future may disclose this relation.
Conclusion
Codon 10 and 25 polymorphism of TGF-β1 gene may be associated with increase susceptibility to psoriasis but did not seem to have a role in the severity of the disease and on serum TGF-β1 level expression.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Michalek IM, Loring B, John SM. A systematic review of worldwide epidemiology of psoriasis. J Eur Acad Dermatol Venereol. 2017;31(2):205–212. doi:10.1111/jdv.13854
2. Lebwohl MG, Bachelez H, Barker J, et al. Patient perspectives in the management of psoriasis: results from the population-based multinational assessment of psoriasis and psoriatic arthritis survey. J Am Acad Dermatol. 2014;70(5):
3. Lebwohl M. Psoriasis. Lancet. 2003;361(9364):1197–1204. doi:10.1016/S0140-6736(03)12954-6
4. Henseler T, Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol. 1985;13(3):450–456. doi:10.1016/S0190-9622(85)70188-0
5. Krueger JG, Krane JF, Carter DM, Gottlieb AB. Role of growth factors, cytokines, and their receptors in the pathogenesis of psoriasis. J Invest Dermatol. 1990;94(6 Suppl):135s–140s. doi:10.1111/1523-1747.ep12876121
6. Lawrence DA. Identification and activation of latent transforming growth factor beta. Methods Enzymol. 1991;198:327–336.
7. Flisiak I, Chodynicka B, Porebski P, Flisiak R. Association between psoriasis severity and transforming growth factor beta(1) and beta (2) in plasma and scales from psoriatic lesions. Cytokine. 2002;19(3):121–125. doi:10.1006/cyto.2002.1953
8. Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor β in human disease. N Engl J Med. 2000;342(18):1350–1358. doi:10.1056/NEJM200005043421807
9. Ince MN, Elliott DE, Setiawan T, et al. Role of T cell TGF-beta signaling in intestinal cytokine responses and helminthic immune modulation. Eur J Immunol. 2009;39(7):1870–1878. doi:10.1002/eji.200838956
10. Massagué J. The transforming growth factor-beta family. Annu Rev Cell Biol. 1990;6:597–641. doi:10.1146/annurev.cb.06.110190.003121
11. Kim SY, Han SW, Kim GW, Lee JM, Kang YM. TGF-beta1 polymorphism determines the progression of joint damage in rheumatoid arthritis. Scand J Rheumatol. 2004;33(6):389–394. doi:10.1080/03009740410010344
12. Grainger DJ, Heathcote K, Chiano M, et al. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet. 1999;8(1):93–97. doi:10.1093/hmg/8.1.93
13. Berndt SI, Huang W-Y, Chatterjee N, et al. Transforming growth factor beta 1 (TGFB1) gene polymorphisms and risk of advanced colorectal adenoma. Carcinogenesis. 2007;28(9):1965–1970. doi:10.1093/carcin/bgm155
14. El-Sherbini SM, Shahen SM, Mosaad YM, Abdelgawad MS, Talaat RM. Gene polymorphism of transforming growth factor-beta1 in Egyptian patients with type 2 diabetes and diabetic nephropathy. Acta Biochim Biophys Sin (Shanghai). 2013;45(4):330–338. doi:10.1093/abbs/gmt003
15. Yamada Y, Miyauchi A, Goto J, et al. Association of a polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to osteoporosis in postmenopausal Japanese women. J Bone Miner Res. 1998;13(10):1569–1576. doi:10.1359/jbmr.1998.13.10.1569
16. Tamizifar B, Lankarani KB, Naeimi S, Rismankar Zadeh M, Taghavi A, Ghaderi A. Promoter polymorphism of transforming growth factor-beta1 gene and ulcerative colitis. World J Gastroenterol. 2008;14(2):243–247. doi:10.3748/wjg.14.243
17. Ohtsuka T, Yamakage A, Yamazaki S. The polymorphism of transforming growth factor-beta1 gene in Japanese patients with systemic sclerosis. Br J Dermatol. 2002;147(3):458–463. doi:10.1046/j.1365-2133.2002.04947.x
18. El-Hadidi HH, Hassan AS, El-Hanafy G, Amr KS, Abdelmesih SF, Abdelhamid MF. Transforming growth factor-β1 gene polymorphism in psoriasis vulgaris. Clin Cosmet Investig Dermatol. 2018;11:415–419. doi:10.2147/CCID.S171403
19. Baran W, Szepietowski JC, Mazur G, Baran E. TGF-β1 gene polymorphism in psoriasis vulgaris. Cytokine. 2007;38(1):8–11. doi:10.1016/j.cyto.2007.04.004
20. Fredriksson T, Pettersson U. Severe psoriasis–oral therapy with a new retinoid. Dermatologica. 1978;157(4):238–244. doi:10.1159/000250839
21. Perrey C, Turner SJ, Pravica V, Howell WM, Hutchinson IV. ARMS-PCR methodologies to determine IL-10, TNF-alpha, TNF-beta and TGF-beta 1 gene polymorphisms. Transpl Immunol. 1999;7(2):127–128. doi:10.1016/S0966-3274(99)80030-6
22. Caca-Biljanovska N, V’Lckova-Laskoska M, Balabanova-Stefanova M, Grivceva-Panovska V. Frequency of delayed-type hypersensitivity to contact allergens in palmo-plantar psoriasis. Prilozi. 2005;26(2):131–141.
23. Paradowska-Gorycka A, Roszak M, Stypinska B, et al. IL-6 and TGF-β gene polymorphisms, their serum levels, as well as HLA profile, in patients with systemic lupus erythematosus. Clin Exp Rheumatol. 2019;37(6):963–975.
24. Di Fusco D, Laudisi F, Dinallo V, et al. Smad7 positively regulates keratinocyte proliferation in psoriasis. Br J Dermatol. 2017;177(6):1633–1643. doi:10.1111/bjd.15703
25. Shull MM, Ormsby I, Kier AB, et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359(6397):693–699. doi:10.1038/359693a0
26. Han G, Williams CA, Salter K, Garl PJ, Li AG, Wang XJ. A role for TGFbeta signaling in the pathogenesis of psoriasis. J Invest Dermatol. 2010;130(2):371–377. doi:10.1038/jid.2009.252
27. Cataldi C, Mari NL, Lozovoy MAB, et al. Proinflammatory and anti-inflammatory cytokine profiles in psoriasis: use as laboratory biomarkers and disease predictors. Inflamm Res. 2019;68(7):557–567. doi:10.1007/s00011-019-01238-8
28. Doi H, Shibata MA, Kiyokane K, Otsuki Y. Downregulation of TGFbeta isoforms and their receptors contributes to keratinocyte hyperproliferation in psoriasis vulgaris. J Dermatol Sci. 2003;33(1):7–16. doi:10.1016/S0923-1811(03)00107-5
29. Zaher H, Shaker OG, EL‐Komy MH, El-Tawdi A, Fawzi M, Kadry D. Serum and tissue expression of transforming growth factor beta 1 in psoriasis. J Eur Acad Dermatol Venereol. 2009;23(4):406–409. doi:10.1111/j.1468-3083.2008.03064.x
30. Flisiak I, Zaniewski P, Chodynicka B. Plasma TGF-beta1, TIMP-1, MMP-1 and IL-18 as a combined biomarker of psoriasis activity. Biomarkers. 2008;13(5):549–556. doi:10.1080/13547500802033300
31. Adbullah TH, Latif II, Ibrahim KK. Role of tumor necrosis factor alpha and transforming growth factor beta as predictive marker for psoriasis patients. Diyala J Med. 2020;19(1):1–7. doi:10.26505/DJM.19015031015
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
