Back to Journals » International Journal of Nanomedicine » Volume 14
Targeted nanoparticle-mediated LHPP for melanoma treatment
Authors Zhang Q, Xiong M, Liu J, Wang S, Du T, Kang T, Liu Y, Cheng H, Huang M, Gou M
Received 29 November 2018
Accepted for publication 12 March 2019
Published 10 May 2019 Volume 2019:14 Pages 3455—3468
DOI https://doi.org/10.2147/IJN.S196374
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Linlin Sun
Qianqian Zhang,1,2,* Meimei Xiong,2,* Jinlu Liu,1,2 Shuai Wang,2 Ting Du,2 Tianyi Kang,2 Yu Liu,2 Hao Cheng,2 Meijuan Huang,1 Maling Gou2
1Department of Thoracic Oncology, West China Hospital, Sichuan University, Chengdu, 610041, People’s Republic of China; 2State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610041, People’s Republic of China
*These authors contributed equally to this work
Background: Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) is a novel tumor suppressor. However, whether LHPP is effective to melanoma has not been investigated. Gene therapy provides a new strategy for the treatment of melanoma. Currently, it suffers from the lack of safe and effective gene delivery systems.
Methods: A CRGDKGPDC peptide (iRGD) modified hybrid monomethoxy poly(ethylene glycol)-poly(D,L-lactide) nanoparticle (iDPP) was prepared and complexed with a LHPP plasmid, forming an iDPP/LHPP nanocomplex. The iDPP/LHPP nanocomplex was characterized by particle size distribution, zeta potential, morphology, cytotoxicity, and transfection efficiency. The antitumor efficacy of the nanocomplex against melanoma was studied both in vitro and in vivo. Further, the potential epigenetic changes in melanoma induced by iDPP/LHPP nanocomplex were evaluated.
Results: The iDPP/LHPP nanocomplex showed high transfection efficiency and low toxicity. Moreover, the nanocomplex displayed a neutral charge that can meet the requirement of intravenous injection for targeted gene therapy. In vitro and in vivo experiments indicated that the iDPP/LHPP nanocomplex significantly inhibited the melanoma growth without causing notable adverse effects. We also found that LHPP played an important role in epigenetics. It regulated the expression of genes related to the proliferation and apoptosis chiefly at the level of transcription.
Conclusion: This work demonstrates that the iDPP nanoparticle-delivered LHPP gene has a potential application in melanoma therapy through regulation of the genes associated with epigenetics.
Keywords: melanoma, LHPP, nanoparticle, gene therapy, epigenetics
Introduction
Melanoma is the deadliest form of skin cancer characterized by rapidly growing and high aggression.1 According to the GLOBOCAN 2018, there will be 287,723 new cases and 60,712 associated deaths worldwide in 2018.2 Traditional therapeutic methods, such as surgery and chemotherapy, have multiple side effects and fail to reduce the recurrence.3,4 Over the past decades, tremendous efforts have been made to develop safer and more effective therapeutics for melanoma.
Epigenetic alterations, including DNA methylation, histone post-translational modifications, and chromatin remodeling, have a crucial role in the regulation of gene transcription, DNA repair, cell apoptosis, and chromosome condensation.5–8 A growing body of evidence indicates that epigenetic alterations contribute to cancer initiation and progression.9,10 Unlike genetic lesions, epigenetic changes can be reversed by epigenetic drugs. In clinical practice, epigenetic drugs have been used alone or in combination with chemo-/radio-therapies to improve survival time for cancer patients.11–13 Protein phosphorylation which holds an important part in epigenetics has been recognized as a powerful way to regulate oncogenic activities.14,15 Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP), a histidine phosphatase, has been reported as a tumor suppressor that can inhibit the proliferation and progression of human hepatocellular carcinoma (HCC).16 However, the antitumor effect of LHPP on melanoma has not been demonstrated. In this regard, we hypothesized LHPP could inhibit the melanoma growth by regulating the expression of genes related to epigenetics.
Gene therapy offers an alternative approach to cancer treatment.17 The application of viral vectors to deliver therapeutic genes into the target cells has made several significant progresses in the field of cancer gene therapy.18 However, the issues of insertion mutagenesis and immunogenicity have limited their applications.19–22 Compared to viral vectors, non-viral vectors are relatively low toxic and immunogenic, but the transfection efficiency is usually inferior.23,24 Nanocarriers are emerging as an important part in non-viral vectors.25,26 Currently, most of the nanocarriers used for gene therapy are cationic nanoparticles that always induce cell necrosis and cause subsequent inflammatory response.27 Furthermore, it would be eliminated by the reticuloendothelial system (RES) after intravenous administration, resulting in low specific targeting ability in vivo.28,29 To overcome this obstacle, non-cationic nanocarriers for safe and effective gene delivery should be explored. Our previous studies have demonstrated that the biodegradable nanoparticles (iDPP), which were prepared by self-assembly of a tumor-targeted peptide (C18-PEG-iRGD), monomethoxy poly(ethlene glycol)-poly(D,L-lactide) (mPEG-PLA), and N-[1-(2,3-dioleoyloxy) propyl]-N,N,N-trimethylammonium chloride (DOTAP), could safely and efficiently deliver genes into melanoma cells.30
In this study, we used the iDPP nanoparticles to deliver the LHPP plasmid for the treatment of melanoma (Scheme 1). The iDPP nanoparticles were electrostatically bound with negatively charged LHPP DNA plasmid, forming a neutral potential gene formulation. This novel nanocomplex is able to meet the requirement of intravenous administration and selective delivery of LHPP gene to the melanoma cells. The potential therapeutic usefulness of iDPP/LHPP nanocomplex was examined in vitro and in vivo. It is expected that the iDPP/LHPP nanocomplex could inhibit the proliferation and induce the apoptosis of melanoma without causing obvious systemic toxicity. Furthermore, the changes in gene expression related to epigenetics were also investigated by real-time polymerase chain reaction (RT-PCR).
Materials and methods
Reagents, cells, and animals
The iRGD peptide (H-[Cys-Arg-Gly-Asp-Lys-Gly-Pro-Asp-Cys]-NH2) was chemically synthesized by GL Biochem (Shanghai, China). Polyoxyethylene stearate (C18-PEG-OH), DOTAP and branched polyethylenimine(PEI,25 kDa) were purchased from Sigma-Aldrich (St Louis, MO, USA). mPEG-PLA (MW 4000) was obtained from Jinan Daigang Biomaterial (Jinan, China). Lipofectamine 2000 was purchased from Invitrogen (USA). Cell cycle detection kit, Annexin V-FITC/PI apoptosis detection kit, and Cell Counting Kit-8 (CCK-8) were obtained from Nanjing KeyGen Biotech. Co. Ltd (Nanjing, China). Caspase-3 activity kit and Bradford protein assay kit were supplied from Beyotime Institute of Biotechnology (Haimen, China).
B16-F10 cells were purchased from the American Type Culture Collection (Rockville, MD, USA) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco).
C57BL/6 (6–8 weeks old) mice were obtained from the Laboratory Animal Center of Sichuan University (Chengdu, China). The procedures of all animal care and the animal experiments were approved and strictly conducted by the Institutional Animal Care and Ethics Committee of Sichuan University (Chengdu, China). The experiments were carried out according to the Institutional Animal Care and Use guidelines of this committee.
Construction and purification of plasmid-based expression vectors: LHPP
The therapeutic LHPP gene was synthesized and inserted into the pVAX plasmid at a Hind III site and an Xba I site, forming the expression plasmid (pLHPP). The empty plasmid pVAX was used as negative control. The recombinant plasmid and negative control plasmid were prepared using the Endofree Plasmid kit (Omega Bio-Tek, Norcross, GA, USA) in accordance with the manufacturer’s instructions and dissolved in sterile endotoxin-free water respectively. The solution was stored at −20 °C for further use.
Preparation and characterization of iDPP/LHPP nanocomplex
To prepare a stable iRGD/DOTAP/mPEG-PLA nanopaticle (iDPP), C18-PEG-iRGD, DOTAP and mPEG-PLA were dissolved respectively in methylene dichloride and mixed in a round bottom flask. Then the above mixture was rotary evaporated to form a transparent film under the condition of 60 °C for 30 min. The film was rehydrated in double-distilled water to create the micelles by self-assembly. The resultant micelles were adjusted to the final concentration of 2 mg/mL. 30
In order to obtain the iDPP/LHPP nanocomplex, iDPP was gently mixed with LHPP plasmid at a mass ratio of 25:1 in DNase-free water, then incubated at room temperature for 30 min. The size distribution and zeta potential of the iDPP and iDPP/LHPP nanocomplex were measured by a dynamic light scattering instrument (Zetasizer, Nano-ZS, Malvern, UK). The equilibration time was 2 min at the test temperature of 25 °C during measurements. All results were presented as the mean of three test runs. The morphologies of the iDPP and iDPP/LHPP nanocomplex were observed under a transmission electron microscope (TEM) (H-600, Hitachi, Japan).
Gel retardation assay
The iDPP/LHPP nanocomplexes with several different mass ratios (0:1, 5:1, 10:1, 15:1, 20:1, 25:1, 30:1) were incubated at room temperature for 30 min. After incubation, they were electrophoresed on 1% (w/v) agarose gel (Thermo Fisher Scientific, U.S.A) stained with Golden View™ for 30 min at 100 V. The electrophoresis gels were detected and photographed using a gel documentation system (Bio-Rad).
Hemolytic tests and erythrocyte aggregation assay
To assess the blood compatibility of iDPP nanoparticles, hemolytic test and erythrocyte aggregation assay were performed. Erythrocytes were isolated from the fresh blood of C57BL/6 mice by centrifuging at 3000 rpm for 10 min. After washing with PBS, the remaining cells were resuspended in saline. For hemolytic tests, the iDPP/LHPP nanocomplexes with different concentrations (0.4, 0.6, 0.8, and 1 mg/mL) were incubated with 2.5 mL of erythrocyte suspension (4%) for 3 h at 37 °C. Saline and distilled water group were used as the controls. Followed by the centrifugation of the mixtures, the supernatants were transferred to a 96-well plate. The degree of hemolysis was assessed by measuring the absorbance at 450 nm using a microplate reader (Bio-Rad 680, USA). For erythrocyte aggregation assay, 200 μL of erythrocyte suspension (2%) was plated in a 24-well plate and then treated with 5 μg of iDPP, iDPP/LHPP, or PEI 25 K for 2 h at 37 °C. Then the erythrocytes were imaged using an optical microscope.
DNase I protection test
To determine the protection of plasmid DNA in iDPP nanoparticles from enzymatic degradation, the DNase I protection test was carried out as previously described.31 Briefly, iDPP/LHPP nanocomplex was treated with 1 U/µL DNase I (Ambion, Austin, TX, USA) in a total volume of 500 μL in 50 mM Tris buffer (pH 7.4) containing 10 mM MgCl2 at 37 °C. After incubation for 0 min, 15 min, 30 min, 1 h, 2 h, 4 h, 4 μL of 250 mM ethylenediamine tetraacetic acid (EDTA) was added to stop DNA degradation. Then 2 μL of Triton X-100 was used to release LHPP from iDPP/LHPP nanocomplex. 5 min later, 4 μL of 10% sodium dodecyl sulfate (SDS) was added at room temperature for 15 min and 10 μL of each sample was analyzed by 1% agarose gel electrophoresis as described above.
In vitro gene transfection
B16-F10 cells were seeded in 6-well plates at a density of 2×105 cells per well in 2 mL of complete medium containing 10% fetal bovine serum (FBS) for 24 h. Thereafter, the medium was replaced with 800 μL of the fresh nonserum medium. A reporter plasmid encoding green fluorescence protein (pGFP, 2 µg per well) was mixed with the materials in serum-free medium. Lipofectamine 2000 was blended with plasmids in Opti-MEM (Gibco). The mass ratios of iDPP/pGFP, PEI25K/pGFP, and Lipofectamine 2000/pGFP were 25/1, 1/1 and 1/1, respectively. VSV-GFP was prepared at a MOI of 0.02. After 6 h of incubation, the medium was replaced by complete DMEM medium and the cells were incubated for additional 24 h. The transfected cells were subsequently observed under a fluorescence microscope (Olympus Corporation, Tokyo, Japan). The expression of green fluorescence protein was quantified by flow cytometry (Calibur, BD, U.S.A).
Cytotoxicity assay
CCK8 assay was used to monitor cell viability after transfection with pGFP (0.08 µg per well). Briefly, cells were plated in 96-well plates at a density of 5×103 cells per well in 100 µL of complete medium containing 10% fetal bovine serum (FBS) and incubated for 24 h. Then cells were treated in triplicate with NS, iDPP/pGFP, PEI25K/pGFP, Lipofectamine 2000/pGFP, and VSV-GFP according to the protocol described above. After incubation for 6 h, the medium was replaced with fresh complete medium. 48 h later, 10 uL of CCK8 reagent was added to each well and incubated for an additional 4 h at 37 °C in the dark. Absorbance value of each well was measured by a microplate reader at a wavelength of 450 nm.
Cellular uptake test
To investigate the cellular uptake efficiency of iDPP/LHPP by B16-F10 cells, pLHPP was labelled with YOYO-1 (Y3601, Thermo Fisher Scientific, U.S.A) according to the manufacturer’s instruction. B16-F10 cells were seeded in 12-well plates at a density of 1.5×105 cells per well and cultured for 24 h. Then YOYO-1-labeled pLHPP was incubated with iDPP, PEI 25K, and Lipofectamine for 30 min, respectively. The cells were transfected with iDPP/LHPP, PEI 25K/LHPP, and lipofectamine/LHPP for 4 h. Next, the cells were collected for the measurement of YOYO-1-derived green fluorescent signal by flow cytometry.
Colony formation assay
One day prior to transfection, B16-F10 cells were seeded into 6-well plates at a density of 1×103 cells per well and cultured in complete medium containing 10% fetal bovine serum (FBS).NS, iDPP/pVAX (50 μg iDPP/2 μg pVAX), iDPP/LHPP (50 μg iDPP/2 μg pLHPP) were prepared in DMEM medium without serum. The cells were treated with those samples separately for 6 h. Then the medium was replaced with 2 mL of DMEM complete medium. 10 days later, the cells were washed with PBS three times and fixed in 4% paraformaldehyde for 20 min. Then the cells were stained with 0.1% crystal violet for observation.
Cell proliferation assay
B16-F10 cells were plated in 96-well plates at a density of 5×103cells per well in 100 uL of DMEM medium and incubated for 24 h. Then the cells were treated with normal saline (NS), iDPP/pVAX nanocomplex (25 μg iDPP/1 μg pVAX) or iDPP/LHPP nanocomplex (25 μg iDPP/1 μg pLHPP) in DMEM medium without serum. After incubation for 6 h, the medium was replaced with complete medium. After transfection for 1, 2, 3 or 4 days, cell viability was evaluated by the CCK8 assay .
Cell cycle detection and apoptosis analysis
B16-F10 cells were seeded into 6-well plates at a density of 2×105 cells per well for 24 h. Then the cells in serum-free medium were treated with NS, iDPP/pVAX (50 μg iDPP/2 μg pVAX), iDPP/LHPP (50 μg iDPP/2 μg pLHPP) separately. After 6 h of incubation, cells were incubated for another 48 h with the replaced 2 mL of DMEM complete medium. The cell cycle status was assessed by flow cytometry using the cell cycle detection kit in accordance with the manufacture’s protocol. Cell apoptosis was examined by flow cytometry using FITC-Annexin V/PI apoptosis detection kit.
Caspase-3 activity assay
After transfection for 36 h, cells in each group were collected and lysed by lysis buffer for 15 min on ice. The proteins in cell lysates were quantified by Bradford assay. The protein concentration in each group was adjusted with pure water to 2 mg/mL. 50 μL of the samples were centrifuged at 16,000 g for 15 min at 4 °C. Then the supernatants were incubated with 40 uL of the reaction buffer and 10 μL of the caspase-3 substrate acetyl-Asp-Glu-Val-Asp p-nitroanilide (Ac-DEVD-pNA) (2 mM). Lysates were incubated at 37 °C for 2 h. Samples were measured with the microplate reader at a wavelength of 405 nm. All the experiments were carried out in triplicates.
Imaging of the activity of the luciferase in vivo
For in vivo gene transfection, pGL6 (Beyotime Biotechnology, China) was used as a luciferase report plasmid. The tumor-bearing mouse model was constructed by subcutaneously injecting 5×105 cells into the female C57BL/6 mice. After 14 days when the tumors have grown to ~1 cm in diameter, the imaging of the luciferase (pGL6) activity was performed. The iDPP/pGL6 nanocomplex (125 μg iDPP/5 μg pGL6) was administrated intravenously.72 h later, the mice were intravenously injected with 150 mg/kg D-luciferin of body weight and anaesthetized by 5% chloralhydrate. 20 min after the substrate injection, the imaging data were collected and monitored under the IVIS Lumina system (Caliper Life Sciences, USA).
Anticancer activity of iDPP/LHPP nanocomplex in vivo
A subcutaneous melanoma tumor model was established on female C57BL/6 mice by subcutaneous injection of B16-F10 cells (about 5×105 cells/100 μL serum-free DMEM) on day 0. Then the mice were randomly divided into three groups (n=5) on day 6 and given the following treatments: NS, iDPP/pVAX nanocomplex (125 μg iDPP/5 μg pVAX) and iDPP/LHPP nanocomplex (125 μg iDPP/5 μg pLHPP). The treatment was performed for five times at an interval of 1 day. The tumor volume of mice was recorded every two days. All mice were sacrificed by cervical vertebra dislocation on day 16. The tumor weight was recorded and analyzed. Tumors and vital organs were gathered and fixed in 4% paraformaldehyde or frozen in liquid nitrogen for future analysis.
Real-time polymerase chain reaction (RT-PCR)
Total RNA was isolated from the treated cells or tumor tissues using an RNA simple total RNA kit (Tiangen, China). cDNA synthesis was performed with total RNA using the Prime Script RT reagent kit (Takara, Tokyo, Japan). The amount of cDNA used as templates was normalized to β-actin. The following sets of primers were used: LHPP, forward: 5′-ATGGCCGCATGGGCTGAG-3′, reverse: 5′-TCACTTGTCCGTGTACTTCAGCA-3′; H2afv, forward: 5′-ATCCACAGACACTTGAAGACTC-3′, reverse: 5′-CATTACCTGCTAACTCCAACAC-3′; EZH2, forward: 5′- AGAGTGGAAGCAGCGGAGGATAC-3′, reverse: 5′-CATTATAGGCACCGAGGCGACTG-3′; CDK8, forward: 5′- GCAACAACCACACTAACGGAACTG-3′, reverse: 5′- GAACGCTGATAGTCGGAGGTCATG-3′; Hdac1, forward: 5′- ACGGCATTGACGACGAATCCTATG-3′, reverse: 5′- CTGAGCCACACTGTAAGACCACTG-3′; caspase3, forward: 5′- GAAACTCTTCATCATTCAGGCC-3′, reverse: 5′- GCGAGTGAGAATGTGCATAAAT-3′; fas, forward: 5′- CAAGTGCAAACCAGACTTCTAC-3′, reverse: 5′- GCACTTTCTTTTCCGGTACTTT-3′; β-actin, forward: 5′-AGAGCTACGAGCTGC CTGAC-3′, reverse: 5′-AGCACTGTGTTGGCGTACAG-3′. Relative quantification of the expression gene was calculated after normalization to β-actin. Experiments were performed in triplicates.
H&E staining and immunohistochemistry
The tumor tissues collected from all groups were fixed in 4% paraformaldehyde solution for more than 48 h, then gradually dehydrated and embedded in paraffin blocks. Tissues embedded in paraffin were sliced into tissue sections of 3–4 µm thickness and stained with hematoxylin and eosin (H&E). Immunohistochemistry for Ki67 was performed using rabbit anti-mouse Ki67 antibody (Novus Biologicals, Littleton, CO, US) according to the previous description. To evaluate the apoptotic cells of tumor tissues, the terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) assay was performed using the In Situ Cell Death Detection kit (Roche Diagnostics Corp11, In, IL, USA) according to the manufacturer’s instructions. Images of all sections were obtained using an optical microscope (Olympus, Japan). The Ki67-positive rate and TUNEL-positive cells were quantified using Image J software.
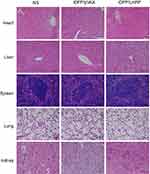
Safety evaluation
To evaluate the safety of iDPP/LHPP nanocomplex, the tumor-bearing mice in each group were continuously observed for the relevant indices such as appearance, physical activity or toxic deaths. The histological sections of vital organs (heart, liver, spleen, lung and kidney) were stained with H&E and observed under the optical microscope (Olympus, Japan).
Statistical analysis
All experimental data were analyzed with GraphPad Prism 6.0 and expressed as the mean ± standard deviation. The decision of statistical significance between two groups was performed by student’s t-test (two-tailed and unpaired). The statistically significant differences were set at *p<0.05, ** p<0.01 and *** p<0.001, respectively.
Results
Preparation and characterization of iDPP/LHPP nanocomplex
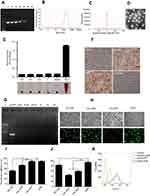
To prepare a nonviral gene delivery system for cancer therapy, we used a tumor targeted peptide C18-PEG-iRGD, mPEG-PLA and DOTAP to self-assemble into a stable iRGD/DOTAP/mPEG-PLA nanopaticle (iDPP). As shown in Figure S1A and B, the mean hydrodynamic particle size and zeta potential of the iDPP nanoparticles were 138±2 nm and 42.37±0.74 mV. Transmission electron microscopy (TEM) images of the iDPP nanoparticle indicated that these particles had a mean size of about 50 nm (Figure S1C). We also investigated the DNA-binding ability of iDPP using gel retardation assay (Figure 1A). When the mass ratio of iDPP to DNA was 25: 1, the anionic DNA was totally retarded. It was confirmed that the iDPP nanoparticle could totally bind the DNA forming a DNA nanocomplex. We further investigated the particle size and zeta potential of iDPP/LHPP nanocomplex. It showed a moderate increase in particle size as 142±2 nm (Figure 1B) without significant difference compared with the iDPP (p=0.39) while a significant decrease as −1.57±0.09 mV in terms of zeta potential (Figure 1C). Morphologic feature of iDPP/LHPP nanocomplex showed that it had a spherical shape with a diameter of about 55 nm (Figure 1D).
To investigate the blood compatibility of iDPP/LHPP nanocomplex, hemolytic test and erythrocyte aggregation assay were carried out. The results indicated that iDPP/LHPP nanocomplex did not induce significant hemolysis (Figure 1E). Furthermore, the erythrocyte aggregation caused by iDPP/LHPP nanocomplex was not observed obviously, while both of PEI25K and iDPP nanoparticles induced significant aggregation of erythrocytes (Figure 1F).
In addition, we evaluated the stability of LHPP encapsulated in iDPP/LHPP nanocomplex against DNase I. DNA was completely degraded within 10 min by DNase I treatment. However, the iDPP nanoparticles protected the LHPP plasmids from enzymatic degradation for 4 h in the presence of DNase I (Figure 1G).
Then we used pGFP as a reporter gene to compare the transfection efficiency and cytotoxicity of iDPP with PEI25K, Lipofectamine 2000 and VSV-GFP (Figure 1H–I). The data revealed that transfection efficiency of iDPP nanoparticles was 88.93±3.43%, which was as high as that of VSV (p=0.810),while PEI25K showed much lower transfection efficiency(38.43±3.52%). In particular, iDPP showed higher transfection efficiency than the commercial agent Lipofectamine 2000 (73.13±4.3%). Further, 24 h after transfection, we used CCK8 assay to measure the viability of transfected cells (Figure 1J). The result showed that delivery of pGFP by PEI25K, Lipofectamine 2000 and VSV-GFP led to cytotoxic effects on B16-F10 cells, while that delivered by iDPP induced little cytotoxicity. Moreover, we performed the cellular uptake experiment to evaluate the tumor targeting ability of the iDPP. As shown in Figure 1K, the B16-F10 cells incubated with YOYO-1 (green) labeled iDPP/LHPP exhibited the highest cellular uptake efficiency among all groups.
Anticancer activity of iDPP/LHPP nanocomplex in vitro
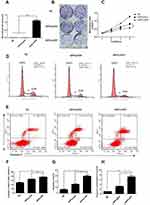
To evaluate the anticancer activity of iDPP/LHPP nanocomplex in vitro, we first performed RT-PCR to confirm the expression of LHPP in B16-F10 cells. pVAX is the plasmid where gene of interest is inserted. After 48 h of transfection, the B16-F10 cells in LHPP treated group expressed high levels of LHPP mRNA while the expressions in other groups were negligible (Figure 2A). We next examined the role of iDPP/LHPP nanocomplex in anti-proliferation and anti-tumorigenesis. The result of colony formation assay indicated that increased expression of LHPP significantly impaired B16-F10 cell proliferation (Figure 2B). CCK8 assay showed a similar result that iDPP/LHPP nanocomplex significantly inhibited the growth of B16-F10 cells compared to other groups (Figure 2C). Next, we used flow cytometry to analyze the cell cycle changes and apoptosis induced by iDPP/LHPP nanocomplex. The cell cycle test showed that iDPP/LHPP nanocomplex caused an arrest of cells in the G0/G1 phase accompanied by a reduction in the number of cells in S phase and G2/M phase, suggesting that the proliferation inhibition effect induced by iDPP/LHPP nanocomplex contributed to G0/G1 cell cycle arrest. (Figure 2D and F). As shown in Figure 2E–G, 29.27±2.38% of apoptotic cells were detected after the treatment with iDPP/LHPP nanocomplex while it was 15.24±1.62% in iDPP/pVAX group and 6.26±1.39% in NS group, respectively. In addition, the activity of caspase-3 was analyzed by measuring the levels of p-nitroaniline (pNA) cleaved from the substrate Ac-DEVD-pNA. As shown in Figure 2H, cells in iDPP/LHPP treatment group showed significant increase in caspase-3 activity. The results implicated that iDPP/LHPP nanocomplex induced apoptosis through the activation of caspase-3 (a common executor of apoptosis). All these results demonstrated that the iDPP/LHPP nanocomplex efficiently expressed LHPP in B16-F10 cells, resulting in the proliferation inhibition and apoptosis induction of melanoma cells.
Anticancer activity of iDPP/LHPP nanocomplex in vivo
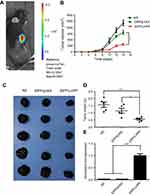
We firstly used pGL-6 as a reporter gene to investigate the tumor targeted ability of the iDPP/pGL-6 nanocomplex in vivo. As shown in Figure 3A, the luciferase accumulated specifically in the tumor tissue after the intravenous administration of iDPP/pGL-6 nanocomplex. The result suggested that iDPP/LHPP nanocomplex would present a high in vivo tumor-targeting efficiency. Next, the antitumor activity of iDPP/LHPP nanocomplex was assessed on the subcutaneous melanoma tumor model. The tumor-bearing mice were divided into three groups and treated with NS, iDPP/pVAX nanocomplex and iDPP/LHPP nanocomplex, respectively. As a result, the tumor growth in iDPP/LHPP treatment group was significantly slower than those in the NS and iDPP/pVAX group (Figure 3B). The representative images of melanoma in each treatment group were shown in Figure 3C. The average tumor weight of mice in iDPP/LHPP treatment group was 0.55±0.15 g, while that in iDPP/pVAX treatment group and NS group were 1.15±0.38 g and 1.56±0.42 g, respectively (Figure 3D). Furthermore, intravenous administration of iDPP/LHPP nanocomplex resulted in enhanced LHPP mRNA expression in the tumor tissues (Figure 3E), suggesting that the iDPP nanoparticles successfully delivered LHPP gene into melanoma cells.
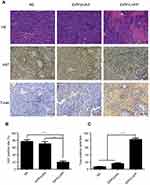
Furthermore, the tumor tissues were analyzed by H&E, immunohistochemistry and TUNEL assay (Figure 4A). The result of H&E assay revealed that iDPP/LHPP treatment group had less tumor cells than other groups. Ki67 immunohistochemistry was performed to study whether the antitumor effect of iDPP/LHPP nanocomplex was associated with inhibition of cell proliferation. Fewer proliferating cells could be observed in the tumor tissues treated with iDPP/LHPP nanocomplex (20±5%) compared to that in iDPP/pVAX treatment group (71.0±10.15%, p<0.01) and NS group (77.67±7.5%, p<0.001) (Figure 4B).TUNEL assay was also conducted to investigate whether the apoptosis was activated by the iDPP/LHPP treatment. As shown in Figure 4C, the number of TUNEL-positive tumor cells in tumor tissues of iDPP/LHPP treatment group significantly increased compared to iDPP/pVAX treatment group and NS group (82.67±6.43 versus 16.0±3.61 or 6.67±2.08, p<0.001).Thus, the results suggested that the anticancer effects of iDPP/LHPP nanocomplex may partly result from the inhibition of proliferation and the induction of apoptosis in tumor cells.
During the animal experiments, we did not find any reduction in physical activity of mice in iDPP/LHPP treatment group. 14 days after inoculation, nearly all mice suffered cachexia except for the mice in iDPP/LHPP treatment group. H&E staining analysis of vital organs (heart, liver, spleen, lung, and kidney) showed that iDPP/LHPP nanocomplex did not cause significant pathological differences (Figure 5).
Epigenetic changes in melanoma induced by iDPP/LHPP nanocomplex
Since we have demonstrated that iDPP/LHPP nanocomplex could inhibit melanoma growth in vitro and in vivo, we then investigated the underlying epigenetic changes in melanoma induced by iDPP/LHPP nanocomplex. Histones are of paramount importance in epigenetics, here we focused on the histone variants and modifications as the important epigenetic factors that may drive the cancer progression. The variant histone H2afv and the histone methyltransferase enzyme EZH2 have been found to have a proliferative effect on melanoma cells.32–34 Moreover, cyclin-dependent kinase 8 (CDK8) deletion could repress EZH2 activity and accelerate tumourigenesis.35 Histone deacetylases (HDACs) which catalyze the removal of acetyl groups from core histones have been shown as a regulator in cancer initiation and progression.36 Among the HDAC family, the histone deacetylase 1 (HDAC1) is believed to regulate most of the observed changes in histone acetylation.37 HDAC1 has been proved to regulate proapoptotic proteins such as caspases 3 and fas.38,39 Thus, RT-PCR analysis was performed to investigate the LHPP regulation of transcriptional expression of these proliferation and apoptosis-related genes in cancer epigenetics. As shown in Figure 6A, overexpression of LHPP downregulated H2afv, EZH2, and CDK8 in the transcriptional state. Furthermore, LHPP could downregulate HDAC1, which may lead to the upregulation of caspase 3 and fas genes (Figure 6B).
Conclusion
In summary, we have developed an iDPP/LHPP nanocomplex which could be administered intravenously for targeted therapy of melanoma. The iDPP/LHPP nanocomplex showed high transfection efficiency and little cytotoxicity. In vitro and in vivo experiments showed that iDPP-mediated delivery of LHPP gene into B16-F10 cells significantly suppressed tumor growth without causing obvious adverse effects. Furthermore, we preliminarily proved that LHPP was able to exert the anti-tumor effect through regulation of the genes associated with epigenetics. To our knowledge, it is the first attempt to apply the iDPP nanoparticle to deliver LHPP gene for the treatment of melanoma. Our results suggested that the iDPP/LHPP gene formulation would have a potential application for melanoma treatment.
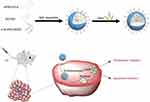 | Scheme 1 Schematic illustration of iDPP/LHPP nanocomplex for the treatment of melanoma. |
Acknowledgments
This work was supported by the National Natural Science Foundation (81572990), Sichuan Science and Technology Program (2019YJ0068), Key Research and Development Projects of Peoples Liberation Army (BWS17J036), and 1·3·5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYYC08007, ZYJC18017).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Li J, Wang Y, Liang R, et al. Recent advances in targeted nanoparticles drug delivery to melanoma. Nanomed Nanotechnol Biol Med. 2015;11(769–794). doi:10.1016/j.nano.2014.11.006
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. doi:10.3322/caac.21492
3. Testori A, Rutkowski P, Marsden J, et al. Surgery and radiotherapy in the treatment of cutaneous melanoma. Ann Oncol. 2009;20(suppl 6):vi22–vi29. doi:10.1093/annonc/mdp257
4. Schadendorf D, van Akkooi ACJ, Berking C, et al. Melanoma. Lancet. 2018;392:971–984. doi:10.1016/s0140-6736(18)31559-9
5. Tough DF, Tak PP, Tarakhovsky A, Prinjha RK. Epigenetic drug discovery: breaking through the immune barrier. Nat Rev Drug Discov. 2016;15(12):835–853. doi:10.1038/nrd.2016.185
6. Mahajan K, Mahajan NP. Cross talk of tyrosine kinases with the DNA damage signaling pathways. Nucleic Acids Res. 2015;43(14):10588–10601. doi:10.1093/nar/gkv1166
7. Hu XT, Kuang Y, Li L, et al. Epigenomic and functional characterization of Junctophilin 3 (JPH3) as a novel tumor suppressor being frequently inactivated by promoter CpG methylation in digestive cancers. Theranostics. 2017;7(7):2150–2163. doi:10.7150/thno.18185
8. Zhang YW, Ghosh AK, Pommier Y. Lasonolide A, a potent and reversible inducer of chromosome condensation. Cell Cycle. 2012;11:4424–4435. doi:10.4161/cc.22768
9. Kanwal R, Gupta S. Epigenetic modifications in cancer. Clin Genet. 2012;81:303–311. doi:10.1111/j.1399-0004.2011.01809.x
10. Mitsiogianni M, Amery T, Franco R, Zoumpourlis V, Pappa A, Panayiotidis MI. From chemo-prevention to epigenetic regulation: the role of isothiocyanates in skin cancer prevention. Pharmacol Ther. 2018;190:187–201. doi:10.1016/j.pharmthera.2018.06.001
11. Moran B, Silva R, Perry AS, Gallagher WM. Epigenetics of malignant melanoma. Semin Cancer Biol. 2018;51(80):80–88. doi:10.1016/j.semcancer.2017.10.006
12. Dawson MA. The cancer epigenome: concepts, challenges, and therapeutic opportunities. Science. 2017;355(1147–1152). doi:10.1126/science.aam7304
13. Pfister SX, Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nat Rev Drug Discov. 2017;16(4):241–263. doi:10.1038/nrd.2016.256
14. Irish JM, Hovland R, Krutzik PO, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118(2):217–228. doi:10.1016/j.cell.2004.06.028
15. Kim E, Kim M, Woo DH, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi:10.1016/j.ccr.2013.04.008
16. Hindupur SK, Colombi M, Fuhs SR, et al. The protein histidine phosphatase LHPP is a tumour suppressor. Nature. 2018;555:678–682. doi:10.1038/nature26140
17. Menezes ME, Talukdar S, Wechman SL, et al. Prospects of Gene Therapy to Treat Melanoma. Adv Cancer Res. 2018;138:213–237. (eds K. D. Tew & P. B. Fisher) Elsevier Academic Press Inc. doi:10.1016/bs.acr.2018.02.007.
18. Wu TL, Zhou DM. Viral delivery for gene therapy against cell movement in cancer. Adv Drug Deliv Rev. 2011;63(8):671–677. doi:10.1016/j.addr.2011.05.005
19. Bouard D, Alazard-Dany N, Cosset FL. Viral vectors: from virology to transgene expression. Br J Pharmacol. 2009;157(2):153–165. doi:10.1038/bjp.2008.349
20. Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–358. doi:10.1038/nrg1066
21. Sokol M, Wabl M, Ruiz IR, Pedersen FS. Novel principles of gamma-retroviral insertional transcription activation in murine leukemia virus-induced end-stage tumors. Retrovirology. 2014;11(1):36. doi:10.1186/1742-4690-11-36
22. Rogers GL, Martino AT, Aslanidi GV, et al. Innate immune responses to AAV vectors. Front Microbiol. 2011;2(10). doi:10.3389/fmicb.2011.00194
23. Yang J, Liu HM, Zhang X. Design, preparation and application of nucleic acid delivery carriers. Biotechnol Adv. 2014;32(4):804–817. doi:10.1016/j.biotechadv.2013.11.004
24. Ramamoorth M, Narvekar A. Non viral vectors in gene therapy- an overview. J Clin Diagn Res. 2015;9:GE1–GE6. doi:10.7860/jcdr/2015/10443.5394
25. Yang CB, Panwar N, Wang Y, et al. Biodegradable charged polyester-based vectors (BCPVs) as an efficient non-viral transfection nanoagent for gene knockdown of the BCR-ABL hybrid oncogene in a human chronic myeloid leukemia cell line. Nanoscale. 2016;8:9405–9416. doi:10.1039/c6nr00996d
26. Liu JB, Song L, Liu S, et al. A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy. Nano Lett. 2018;18:3328–3334. doi:10.1021/acs.nanolett.7b04812
27. Wei XW, Shao B, He Z, et al. Cationic nanocarriers induce cell necrosis through impairment of Na+/K+-ATPase and cause subsequent inflammatory response. Cell Res. 2015;25(2):237–253. doi:10.1038/cr.2015.9
28. Romberg B, Hennink WE, Storm G. Sheddable coatings for long-circulating nanoparticles. Pharm Res. 2008;25:55–71. doi:10.1007/s11095-007-9348-7
29. Jones CH, Chen CK, Ravikrishnan A, Rane S, Pfeifer BA. Overcoming nonviral gene delivery barriers: perspective and future. Mol Pharm. 2013;10:4082–4098. doi:10.1021/mp400467x
30. Luo L, Yang Y, Du T, et al. Targeted nanoparticle-mediated gene therapy mimics oncolytic virus for effective melanoma treatment. Adv Funct Mater. 2018;28(12). doi:10.1002/adfm.201800173
31. He ZY, Wei XW, Luo M, et al. Folate-linked lipoplexes for short hairpin RNA targeting claudin-3 delivery in ovarian cancer xenografts. J Control Release. 2013;172(3):679–689. doi:10.1016/j.jconrel.2013.10.015
32. Kim KH, Roberts CWM. Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–134. doi:10.1038/nm.4036
33. Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106(2):243–247. doi:10.1038/bjc.2011.551
34. Vardabasso C, Hake SB, Bernstein E. Histone variant H2A.Z.2: a novel driver of melanoma progression. Mol Cell Oncol. 2016;3(2). doi:10.1080/23723556.2015.1073417
35. McCleland ML, Soukup TM, Liu SD, et al. Cdk8 deletion in the Apc(Min) murine tumour model represses EZH2 activity and accelerates tumourigenesis. J Pathol. 2015;237:508–519. doi:10.1002/path.4596
36.
37. Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi:10.1146/annurev.pharmtox.45.120403.095825
38. Zhang XD, Gillespie SK, Borrow JM, Hersey P. The histone deacetylase inhibitor suberic bishydroxamate regulates the expression of multiple apoptotic mediators and induces mitochondria-dependent apoptosis of melanoma cells. Mol Cancer Ther. 2004;3:425–435.
39. Cacan E. Histone deacetylase-1-mediated suppression of FAS in chemoresistant ovarian cancer cells. Anticancer Res. 2016;36:2819–2826.
Supplementary material
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.