Back to Journals » Nanotechnology, Science and Applications » Volume 12
Static DNA Nanostructures For Cancer Theranostics: Recent Progress In Design And Applications
Authors Jahanban-Esfahlan R, Seidi K, Jahanban-Esfahlan A , Jaymand M, Alizadeh E, Majdi H, Najjar R , Javaheri T , Zare P
Received 14 August 2019
Accepted for publication 13 September 2019
Published 15 October 2019 Volume 2019:12 Pages 25—46
DOI https://doi.org/10.2147/NSA.S227193
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Israel (Rudi) Rubinstein
Rana Jahanban-Esfahlan,1,2 Khaled Seidi,3 Ali Jahanban-Esfahlan,4 Mehdi Jaymand,5 Effat Alizadeh,1 Hasan Majdi,6 Reza Najjar,7 Tahereh Javaheri,8 Peyman Zare9
1Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz 9841, Iran; 2Student Research Committee, Tabriz University of Medical Sciences, Tabriz 9841, Iran; 3Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz 9841, Iran; 4Stem Cell Research Center, Tabriz University of Medical Sciences, Tabriz 9841, Iran; 5Nano Drug Delivery Research Center (NDDRC), Kermanshah University of Medical Sciences, Kermanshah 9883, Iran; 6Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz 9841, Iran; 7Polymer Research Laboratory, Faculty of Chemistry, University of Tabriz, Tabriz 9841, Iran; 8Ludwig Boltzmann Institute for Cancer Research, Vienna 1090, Austria; 9Faculty of Medicine, Cardinal Stefan Wyszyński University in Warsaw, Warsaw 01-938, Poland
Correspondence: Tahereh Javaheri
Ludwig Boltzmann Institute for Cancer Research, Institute of Animal Breeding and Genetics, University of Veterinary Medicine, Vienna, Austria
Tel +43-1-25077 563
Fax +43-1-40160 931300
Email [email protected]
Peyman Zare
Faculty of Medicine, Cardinal Stefan Wyszyński University in Warsaw, Warsaw 01-938, Poland
Tel +48884842847
Email [email protected]
Abstract: Among the various nano/biomaterials used in cancer treatment, the beauty and benefits of DNA nanocomposites are outstanding. The specificity and programmability of the base pairing of DNA strands, together with their ability to conjugate with different types of functionalities have realized unsurpassed potential for the production of two- and three-dimensional nano-sized structures in any shape, size, surface chemistry and functionality. This review aims to provide an insight into the diversity of static DNA nanodevices, including DNA origami, DNA polyhedra, DNA origami arrays and bioreactors, DNA nanoswitch, DNA nanoflower, hydrogel and dendrimer as young but promising platforms for cancer theranostics. The utility and potential of the individual formats in biomedical science and especially in cancer therapy will be discussed.
Keywords: static DNA nanostructures, cancer treatment, biosensing
Introduction
The integration of nanotechnology into other biomedical sciences has been a blessing for all researchers around the world. Various nanocarriers including polymeric nanoparticles such as hydrogeles, dendrimers, polymerosomes, lipid nanocarriers such as liposomes, micelles, organic nanoparticles such as carbon nanotubes, fullerenes, and non-organic nanoparticulates of gold, silver, magnetic Fe3O4, etc. have often been used for various purposes in the detection and cure of human diseases, in particular, cancer.1–3 Nevertheless, various physiological/physical barriers still need to be addressed in relation to: (i) tumor cell heterogeneity such as clonal evolution and alteration/loss of tumor cell-specific surface markers, (ii) microenvironment of cancer cell such as the presence of a dense extracellular matrix, poor vascular system, low pH, etc., (iii) presence of different physiological barriers such as plasma/nuclear/organelle membranes, blood-brain barrier (BBB), etc; (iv) properties of nanoparticles, including their safety profile, stability, drug encapsulation efficiency, etc.4,5 These barriers have all directed experiments towards the production of the ideal nanoparticle capable of sensing, adapting and reacting to the surrounding microenvironment.
Formulation of such an intelligent and adaptable nanodevice requires a high degree of programmability, a potential, which is hopefully presented by Watson-Crick base pairing principle defined for nucleic acid hybridization. In the first steps of imagining such a fantasy, Seeman used branched DNA with sticky ends to prepare and introduce the first two-dimensional (2D) DNA crystal structure in 2003.6 One year later, Turberfield et al, attempted the one-stage self-assembly of the first DNA cage (DNA tetrahedra) as a typical example of DNA-fabricated nanocarrier (DNA box) for encapsulation of biomolecules.7 Later, functionalization strategies, such as the integration of aptamers, allowed construction of more advanced DNA boxes with the controllable opening-closing performance of box/lid to allow for demand-driven freight exposure/release (box opening) for simultaneous biosensing and the triggering of a therapeutic effect only at the desired site (cancer cells). In addition, due to the unique sequence of DNA molecule used in the construction of these finite-sized DNs, the resulting structures are fully addressable and adaptable in terms of conjugation with a variety of functional units such as small molecules, macromolecules and inorganic nanoparticles that can be easily and accurately (with nanometer precision) placed anywhere on or in such structures (Table 1).
 |  |  |  |
Table 1 Potential Of DNA Nanostructures In Biomedicine |
DNA objects can fall in two categories, as static and dynamic formats. While dynamic formats such as walkers, tweezers, rotor/motor, etc. are capable of cycling movements and stepping/walking on defined DNA tracks using energy derived from DNA strand displacement, the use of static formats is widely accepted and furthur advanced in the context of cancer theranostics and nano drug delivery vehicles for cancer monitoring and treatment. Accordingly, this review aims to provide an overview of the current design formats, benefits and applications of static DNA nanostructures (DNs) including DNA origami nanostructures (DONs), DNA arrays/bioreactors, DNA polyhedra (DNA cage), DNA nanoflower, hydrogel, dendrimer and DNA nanoswitch as emerging promising platforms for cancer treatment in the preclinical settings.
Static DNA Structures For Cancer Theranostics
DNA nanostructures have been extensively used for the study and manipulation of biological processes, which is critical to understand the molecular mechanism underlying pathological/disease-related condition in biomedicine. DN has also found numerous applications in the field of biosensing, detection, treatment, and drug/gene delivery of various cargoes, including nucleic acids, proteins, aptamers, siRNA, miRNAs, and nanoparticles. Accordingly, functionalized DNA structures with theranostic targeting moieties were used for the simultaneous mapping and targeting of various genetic, immunological, infectious, cancer and metabolic diseases. (see Table 1).8 In this section, we will explain the use of static DNs in biomedicine and oncology practice.
DNA Origami
As a key method for the production of DN, DNA origami objects can be adapted in both static and dynamic forms. Looking at the methods for production of DN, there are the scaffold-free tile assembly method (Figure 1A) and the scaffold DNA origami method (Figure 1B). The multi-strand tile method is a bottom-up approach used in the first preparation of two-directional DNA polyhedra. In short, in this process, each tile is made from several single stranded DNA (ssDNA) with sticky ends, where cohesion between complementary strands from different tiles can lead to the assembly of larger structures such as 2D arrays as well as polyhedra (Figure 1A). In the meantime, DNA origami follows a top-down scaffold-mediated approach in which a long ssDNA usually M13 genomic DNA is forced to fold into DONs using short ssDNA oligonucleotides called staples. What’s more, staple act as docking sites for functionalization by nanoparticles and biomolecules. In this way, and with the help of computer programs DNA origami objects can be designed and assembled in almost any form.99 These modifications together with biocompatibility and biostability100–102 have made DNA origami nanostructures suitable platforms for targeted delivery, imaging, and controlled release of therapeutic cargoes.14,103–107
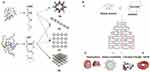 |
Figure 1 Concept of DNA tile and DNA origami assembly methods. (A) Tile method for mounting DNA grids. Reprinted with permission from Manuguerra I, Grossi G, Thomsen RP, et al. Construction of a polyhedral DNA 12-arm junction for self-assembly of wireframe DNA lattices. ACS Nano. 2017;11(9):9041–9047. doi:10.1021/ acsnano.7b03538.165 Copyright (2017) American Chemical Society. (B) a, Principle design of DNA origami. Adapted by permission from Springer Nature: Nature, Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. 2006;440:297. doi:10.1038/nature04586.166 Copyright (2006). b,d, DNA origami objects. b, From Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332(6027):342–346. doi:10.1126/science.1202998.167 Reprinted with permission from AAAS. c, DNA origami pattern. Reprinted by permission from Nature Springer: Nature Nanothechnology, Zhang F, Jiang S, Wu S, et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. 2015;10:779. doi:10.1038/nnano.2015.162.168 Copyright (2015). d, From Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325(5941):725–730. doi:10.1126/science.1174251.169 Reprinted with permission from AAAS. |
DONs are functional as immune-activating programmable adjuvants.33 They are also efficient as simultaneous dual-modality bioimaging and cancer targeting nanotools in vivo14 (Figure 2A). In addition, DONs can be used to enhance the efficacy of chemotherapy (Figure 2B),108 reduce drug side effects, and bypass drug resistance.19,30 Notably, DONs with a different twist allow tuning drug release kinetics of the DNA intercalating Doxorubicin (Dox) for optimum delivery to tumor cells (Figure 2C).21,107 Recently, DNA origami ultrasensitive biosensors namely DNA origami nanoantennas have been successfully adapted for biosensing and superresolution microscopy of Zika-virus nucleic acids at the single molecule level.109
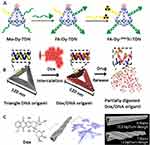 |
Figure 2 DON for targeted drug delivery. (A) Multiple-armed tetrahedral DNA nanostructure (TDNs) for dual-modality in vivo imaging and targeted cancer therapy. Adapted with permission from Jiang D, Sun Y, Li J, et al. Multiple-armed tetrahedral DNA nanostructures for tumor-targeting, dual-modality in vivo imaging. ACS Appl Mater Interfaces. 2016;8(7):4378–4384. doi:10.1021/ acsami.5b10792.14 Copyright (2016) American Chemical Society. (B) Triangle-shaped DNA origami affords optimum drug internalization for cancer chemotherapy. Adapted with permission from Zhang Q, Jiang Q, Li N, et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano. 2014;8(7):6633– 6643. doi:10.1021/nn502058j.108 Copyright (2014) American Chemical Society. (C) DON with a different degree of twist and relaxation achieves tunable drug release kinetics. Adapted with approval from Zhao YX, Shaw A, Zeng X, Benson E, Nystrom AM, Hogberg B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano. 2012;6(10):8684–8691. doi:10.1021/nn3022662.21 Copyright ACS, https://pubs.acs.org/doi/abs/10.1021%2Fnn3022662. Further permissions related to the material excerpted should be directed to ACS. |
More recently, DONs have been shown to be useful tools for investigating antigen-antibody interactions to determine the strongest binding to antibodies in the immune system affecting antibody development for cancer immunotherapy and the vaccine development. In this way, DNA origami was used to represent nanoscale patterns of antigens. This is the first report to provide detailed information on how the antigen distances ranging from 3 to 17 nm can affect the binding affinity of mono and bivalent antibodies, as they have been shown to bind strongly to two antigens when a preferable 16 nm antigenic distance is reached.110 Similarly, DNA origami nanoarrays can interrogate the clustering behavior of cell-surface receptors, because DN allows the representation of receptor ligands in an extremely customizable manner, with adjustable factors such as ligand number, spatial localization and, multivalency.111 As proof of concept, they are used to study epidermal growth factor (EGF) and integrin signaling with nanoscale spatial resolution. That, using multivalent DNA biochips revealed cooperative work of EGF-integrin in modulating the behavior of melanoma cancer cell in a number and ratio-dependent manner.112 Furthur examples of DON in cancer therapy and drug delivery can be found in.99,113
DNA Cage: Polyhedra
DNA polyhedra has proven to be a promising class of drug-carriers frequently used for biomedical applications. The advantage of polyhedra compared over other structures such as DNA origami is their small size and cost efficiency. One of the most popular structures in this class is the tetrahedron introduced by Turberfield.114 This specific example can be constructed from only four strands with near-quantitative yields within 30 seconds and is compression-stable enough to be imaged by atomic force microscopy.
Polyhedra represents a porous structure and is suitable for chemical modifications. In addition, it can be reconfigured with a high load capacity by external triggers for on-command drug release. Numerous applications in multi-modality medical diagnostics and treatments (Figure 3) are proposed for these structures.115 DNA polyhedra can be filled with one or more nanoparticles (Figure 3A),20,93 proteins (eg, cytochrome C (Figure 3B),116 β-galactosidase,117 catabolite activator protein (CAP),83 nucleic acids (eg, siRNA),50 or other biomolecules118 to enable their passive or active on-target delivery. For example, encapsulation affords conditional activation of caged enzymes/transcription factors upon exposure to their specific protein/DNA substrates.83 Alternatively, the device protects its cargo (eg, siRNA) from nuclease degradation and site-specific cleavage.50
 |
Figure 3 DNA nanocage. (A) Inorganic nanoparticle inside the DNA cage as a hybrid drug delivery system. Reprinted with permission from Zhang C, Li X, Tian C, et al. DNA nanocages swallow gold nanoparticles (AuNPs) to form AuNP@DNA cage core-shell structures. ACS Nano. 2014;8(2):1130–1135. doi:10.1021/nn406039p.20 Copyright (2014) American Chemical Society. (B) Single protein encapsulated in rigid DNA tetrahedron nanocage. Reprinted with permission from Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew Chem Int Ed Engl. 2006;45(44):7414–7417. doi:10.1002/anie.200603392. John Wiley and Sons.116 (C) Doxorubicin-loaded tumor-penetrating peptide-modified DNA tetrahedron. Reprinted with permission from Xia Z, Wang P, Liu X, et al. Tumor-penetrating peptide-modified DNA tetrahedron for targeting drug delivery. Biochemistry. 2016;55 (9):1326–1331. doi:10.1021/acs.biochem.5b01181.122 Copyright (2016) American Chemical Society. (D) DNA nanosuitcases for encapsulation and conditional release of siRNA. Reprinted with permission from Bujold KE, Hsu JCC, Sleiman HF, Optimized DNA. “Nanosuitcases” for encapsulation and conditional release of siRNA. J Am Chem Soc. 2016;138(42):14030–14038. doi:10.1021/jacs.6b08369.50 Copyright (2016) American Chemical Society. (E) DNA tetrahedron structured probe (TSP) for protein biosensing. Adapted with permission from Pei H, Lu N, Wen Y, et al. A DNA nanostructure-based biomolecular probe carrier platform for electrochemical biosensing. Adv Mater. 2010;22(42):4754–4758. doi:10.1002/adma.201002767. John Wiley and Sons.126 |
The surface modification of DNA polyhedra with disease-specific markers (eg, folate or oxidized low-density lipoprotein receptor-1” (LOX-1)) not only leads to active targeting but also to more than 30-40-fold internalization efficiency.119,120 DNs can encapsulate toxic and hydrophobic agents, such as Dox for efficient chemotherapy of cancer cells while reducing damage to normal cells.121 The decoration of the Dox-encapsulated DNA cage with tumor-penetrating peptide increases the cellular uptake of DNA tetrahedron for the targeted administration of toxic chemotherapeutic in vitro (Figure 3C).122 Dox-loaded DNA nanoblocks are found in the endocytic vesicles after entry into the target cells, where the acidic milieu triggers the release of drugs from these DNA nanostructures and induces selective augmented cytotoxic actions of Dox (>40 times) on folate receptor expressing cancer cells, while promoting the degradation of nanostructures that prevents their accumulation in the cells.120
DNA tetrahedra are examples of DNA nanocages for large molecules or as building blocks for more complex nano-devices enabling rapid and reliable assembly with quantitative yields, in vitro and in vivo structural stability, and plasticity.7,114,123 As proof of concept, DNA tetrahedra that can easily penetrate mammalian cells have been shown to be stable in cytoplasm.59 A recent report also showed that multivalent DNA tetrahedrons functionalized with CpG motifs are permeable to cells and stimulate an immune response with applications in vaccine development and immunotherapy.35 SiRNA administration using DNA tetrahedra was reported as a vehicle for targeted gene therapy. The adaptability of the design permits precise control of nanoparticle size, the spatial orientation, and density of targeting ligands on the nanoparticle surface or the inside, which could lead to enhanced delivery of the siRNAs into cells and efficient gene silencing.124 Moreover, DNA polyhedra were used as carriers of chemotherapeutics to overcome multi-drug resistance in cancer cells.118 Similarly, aptamer-decorated DNA icosahedral nanoparticles were used to deliver Dox molecules.58 The construction of cage-like DNA structures called nanosuitcases was reported for siRNA loading and on-demand release upon recognition of an oligonucleotide marker (eg, mRNA or a microRNA (miRNA)(Figure 3D).50 In addition, the pH-triggered controlled release of Dox-intercalated nanoring functionalized with mucin aptamer (MUC1) was shown to have therapeutic potential for breast cancer.125 In another example, hybrid systems such as DNA polyhedron-caged gold nanoparticles (AuNP) were produced for the controlled release of Dox when triggered by pH and DNA enzyme (T7 exonuclease).93 Moreover, DNA tetrahedron structures are used as molecular probes with high-sensitivity for DNA target detection and single nucleotide polymorphism (SNP) typing of biological fluids, as well as protein biosensing (Figure 3E) in vitro.126
DNA Arrays And Bioreactors
Another advantage of DNA origami which deserves to be discussed in a separate section, is its potential for the fabrication of DNA arrays/ bioreactors, which can be achieved by site-specific functionalization of DNA origami structures with proteins, nanoparticles, antibodies, enzymes, etc. (Figure 4) with well-defined regularity and periodicity is of great interest to the scientific community.127 Applications include sensing, catalysis, and highly ordered device fabrication.128 The addressability of DNA origami structures enables site-specific localization of several functional groups on their surfaces. Functionality could be achieved by i) covalently bound chemical groups (eg, amine, thiol, and carboxyl groups) or molecules (eg, biotin), or ii) DNA overhangs (eg, aptamers).129 In one of these systems, the chemical conjugation of amino-modified DNA oligonucleotides was demonstrated with chlorohexane or benzylguanine groups as orthogonal tags for the genetic modification of proteins of interest fused with Halo-Tag or Snap-tag, respectively. Authors reported orthogonal modified DNA origami by decorating DNA origami with multiple proteins including mKate-Snap, CCP-Halo, and mSTV, which resemble the characteristic of human face including mouth, eyes, and nose, respectively (Figure 4A).130 Another study used DNA-binding potential of zinc finger proteins for site-specific protein positioning on DNA origami.131 Previous formats of DNA nanoarrays were also attempted in 2005 by DNA hybridization-mediated adhesion of Au nanoparticles of different sizes to prescribed locations on DNA tiles, producing alternating rows of the two components (Figure 4B).132 DNA nanotubes conjugated with two enzymes, horseradish peroxidase, and glucose oxidase, demonstrated enzymatic cascade, as an example of the nanoscale DNA-based bioreactor.127 Moreover, punched DNA origami assembly was exploited for the design of robust and precisely programmed 2D streptavidin nanoarrays by the periodical embedding of biotinylated nanowells in DNA origami structures (Figure 4C).133
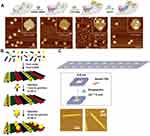 |
Figure 4 DNA origami arrays and bioreactors. (A) multi-protein decoration of DNA origami structures (arrays) resembling a human face. Adapted with permission from Sacca B, Meyer R, Erkelenz M, et al. Orthogonal protein decoration of DNA origami. Angew Chem Int Ed Engl. 2010;49(49):9378–9383. doi:10.1002/anie.201005931. John Wiley and Sons.130 (B) Assembly steps for the 2D nanocomponent arrays. Adapted with permission from Pinto YY, Le JD, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Sequence-encoded self-assembly of multiple-nanocomponent arrays by 2D DNA scaffolding. Nano Lett. 2005;5(12):2399–2402. doi:10.1021/nl0515495.132 Copyright (2005) American Chemical Society. (C) 2D streptavidin nanoarrays on rectangular DNA origami surface. Adapted with permission from Kuzuya A, Kimura M, Numajiri K, et al. Precisely programmed and robust 2D streptavidin nanoarrays by using periodical nanometer-scale wells embedded in DNA origami assembly. Chembiochem. 2009;10(11):1811–1815. doi:10.1002/cbic.200900229. John Wiley and Sons.133 |
DNA Nanoflowers
Long, single-stranded DNA and pyrophosphate released from (in vitro) polymerase reactions can form dense, compact organic-inorganic composite nanomaterials, known as nanoflowers (NFs).134 In contrast to the key role of DNA base pairing in DNA nanostructures formation, NF assembly is independent of DNA hybridization. In addition, unlike other DNs, the shape of nanoflowers cannot be precisely programmed (Figure 5).135
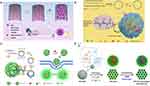 |
Figure 5 DNA nanoflower preparation and its biomedical applications. (A) miRNA initiated the growth and blooming of DNA nanoflower in nanochannel. Adapted with permission from Shi L, Mu C, Gao T, et al. DNA nanoflower blooms in nanochannels: a new strategy for miRNA detection. Chem Commun. 2018;54(81):11391–11394. doi:10.1039/c8cc05690k. Permission conveyed through Copyright Clearance Center, Inc.135 (B) DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Adapted from Hu R, Zhang X, Zhao Z, et al. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew Chem Int Ed Engl. 2014;53(23):5821–5826. doi:10.1002/anie.201400323. John Wiley and Sons.10 (C) RCR-based assembly of NFs loaded with chemotherapeutics for targeted drug delivery. Reprinted by permission from Springer Nature: Lv Y, Hu R, Zhu G, et al. Preparation and biomedical applications of programmable and multifunctional DNA nanoflowers. Nat Protoc. 2015;10(10):1508–1524. doi:10.1038/nprot.2015.078.136 Copyright (2015). (D) Synergistic immunotherapy of cancer by DNA-RNA microflowers (iDR-MFs) loaded with tumor neoantigens. Adapted with permission from Zhu G, Mei L, Vishwasrao HD, et al. Intertwining DNA-RNA nanocapsules loaded with tumor neoantigens as synergistic nano-vaccines for cancer immunotherapy. Nat Commun. 2017;8(1):1482. doi:10.1038/s41467-017-01386.68 |
NF can be assembled using DNA building blocks by a PCR reaction such as rolling circle amplification (RCA). This reaction requires only a primer and template DNA to control over size, construct and functionalization of nanoflowers. This means that the rational design of any RCR-suitable templates allows the elongated DNA building blocks to carry a large number of concatemer functional or structural moieties, which further condense into NFs.136 Due to the dense DNA packaging, NFs resist to nuclease degradation, dissociation or denaturation. Recently, the NF blooming (formation) principle has been used to design a simple and sensitive electrochemical biosensing system. In this preparation, the NF blooming in the nanochannels has been tailored to occur upon binding of target miRNA, followed by binding of a circle template DNA and a specific primer to the captured miRNA, triggering RCA and NF blooming in the nanochannels. NF formation increased steric hindrance in the channels, which reduced the anodic current of potassium ferricyanide (K3[Fe(CN)6]), thus producing enhanced electrochemical detection signals (Figure 5A).135
In addition to excellent biostability, NFs exhibit excellent photostability. In this sense, construction of aptamer conjugated multicolor fluorescence resonance energy transfer (FRET)-NFs is reported with applications in multiplexed cellular bioimaging and drug screening, in which NF were integrated with three different fluorophores (FAM, Cy3, and ROX), therapeutic drug payloads (Dox), and two aptamers (sgc8 and MUC 1). In addition, single wavelength excitation leads to multi-fluorescence emissions of NF, allowing monitoring of drug transport for targeted drug delivery (Figure 5B).10 Moreover, immuno-nanoflowers have been developed for nuclease resistance and efficient CpG delivery.36 CpG NFs were found to be potent immunostimulators capable of triggering the proliferation of macrophage-like cells that stimulated the secretion of immunostimulatory cytokines that induced apoptosis and necrosis of cancer cells. Moreover, pH-responsive multifunctional DNA NFs containing cancer-targeting aptamer, fluorophores, and Dox molecules were used to prevent drug efflux and improve drug retention in MDR cells, thus bypassing MDR and reducing side effects (Figure 5C).136,137 In addition, using microflower nanosystem, synergistic nanovaccines were constructed for effective immunotherapy of cancer. The NFs were formulated to comprise intertwining DNA-RNA nanocapsules (iDR-NCs) conjugated with DNA CpG, STAT3 short hairpin RNA (shRNA) adjuvants, as well as tumor-specific peptide neoantigens. shRNA in iDR-NCs synergistically activated antigen-presenting cells (APCs), primed CD8+ T cells, induced T cell memory, and markedly suppressed the progression of neoantigen-specific colorectal tumors (Figure 5B).68
In addition, water remediation –using magnetite nanoparticles that absorb and remove heavy metals– can be achieved by hybrid nano-flowers consisting of single-crystalline petals coated with magnetite nanoparticles. Additionally, hybrid nanoflowers (hNFs) consisting of organic (enzymes)-inorganic (metal ions) particles is reported to enhance catalytic activities and stability of proteins under various experimental conditions.138,139
DNA Dendrimers
As relatively complicated DNs, DNA dendrimer with a branched configuration can be developed by the facile modular assembly. DNA dendrimers are layer-by-layer self-assembled functional branched DNA units (eg, Y-shaped building blocks) made through predesigned base-pairing hybridization. These somewhat bulkier DNs form promising scaffolds for versatile biomedical applications (Figure 6). For example, they can accommodate more docking sites for loading drugs or other functional materials. Moreover, by implementing functional DNA units, these structures can be constructed into targeted and stimuli-responsive drug delivery systems. Also, sterically crowded conformations, homogeneous controlled sizes, and structural stability are additional advantages of dendrimeric nanostructures. Furthermore, they can be used as biosensing devices. For instance, DNA dendrimers coupled with DNA nanomachines (DNA circuits) has realized an ultrasensitive biosensor with a detection limit of 0.0661 pg/mL and 0.062 nM for laminin15 and KRAS gene fragment (Figure 6A),140 respectively. Tailoring the capture and reporter DNA/protein units can extend the prognostic value of such dendrimer-based self-amplification system for detection of a different disease-related species with high sensitivity in biological samples. Moreover, achieving a therapeutic effect, catalytically self-assembled siRNA-based gene silencing via functionalized DNA dendritic complexes was constructed. Incorporated sgc8c aptamers to DNA dendrimers enabled targeting of overexpressed protein tyrosine kinase-7 (PTK7) receptor on tumor cells and led to higher gene silencing efficiency with lower cytotoxicity when compared to the commercial cationic lipid transfection agents.141 Another study demonstrated that the formulation of dendrimeric siRNA improves condensation, stability, and gene silencing efficiencies.52 Moreover, branched DNA constructs –containing anti-miRNA– have been used for oncomiRNA targeting of cancer. In another report, a multifunctional dendrimeric-based system composing of tumor-targeting ligands –such as MUC1, AS1411, and ATP aptamers– was used for high loading and controlled delivery of an anthracycline drug called “epirubicin” to cancer cells. MUC1 and AS1411 aptamer were coated on the surface of dendrimer to assist crossing the cellular and nuclear membranes, respectively. Meanwhile, ATP aptamer incorporated in the building blocks directed dendrimer to the ATP-enriched lysozymes where more dendrimer disassembly can occur.28 Furthermore, ligase-independent efficient self-assembly of DNA dendrimer is reported by annealing DNA units with elongated adhesive ends. These structures were used for delivery of immunostimulatory CpG DNA encoding tumor necrosis factor-α to immune cells.142 Programmable DNA dendrimers coated with CpG-containing hairpin-loops were found to trigger stronger immune responses than those conjugated with linear CpG. Further surface functionalization of DNA dendrimer with TAT –a classic cell-penetrating peptide– enhanced cell internalization and cytokines production (Figure 6B).34
 |
Figure 6 Design and application of DNA dendrimer in biomedicine. (A) DNA dendrimer–streptavidin for efficient signal amplification for biosensing. Adapted with permission from Zhao Y, Hu S, Wang H, et al. DNA dendrimer–streptavidin nanocomplex: an efficient signal amplifier for construction of biosensing platforms. Anal Chem. 2017;89(12):6907–6914. doi:10.1021/acs. analchem.7b01551.140 Copyright (2017) American Chemical Society. (B) Multifunctional DNA dendrimers. Adapted with permission from Qu Y, Yang J, Zhan P, et al. Self-assembled DNA dendrimer nanoparticle for efficient delivery of immunostimulatory CpG motifs. ACS Appl Mater Interfaces. 2017;9(24):20324–20329. doi:10.1021/acsami.7b05890.34 Copyright (2017) American Chemical Society. |
Possessing a similar mesh-like structure formed out of branched building blocks, DNA hydrogels with even broader biomedical as well as bioelectronic applications are developed, which we discuss in the next chapter.
DNA Hydrogel
A further advantage of DNA as a natural polymer is its capability to act as anirreplaceable building block for the construction of swollen networks of cross-linked DNA in an aqueous solution called DNA hydrogels.143 DNA can be the only component of a hydrogel, the backbone or cross-linker, which connects the main building blocks (hybrid hydrogels) through physical entanglement or chemical reactions (Figure 7).144,145
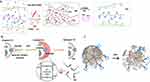 |
Figure 7 Design and application of DNA hydrogels in biomedicine. (A) Schematic representation of the photo-responsive properties of DNA hydrogels for on-demand cargo release of the payloads. Adapted with permission from Kang H, Liu H, Zhang X, et al. Photoresponsive DNA-cross-linked hydrogels for controllable release and cancer therapy. Langmuir. 2011;27(1):399–408. doi:10.1021/la1037553.146 Copyright (2011) American Chemical Society. (B) Apatmer (EpCAM) binding-mediated catalysis of HCR control gel-sol state of ATP-responsive DNA hydrogel, resulting in cloaking (gel state) and releasing (sol state) of circulating tumor cells. Adapted with permission from Song P, Ye D, Zuo X, et al. DNA hydrogel with aptamer-toeholdbased recognition, cloaking, and decloaking of circulating tumor cells for live cell analysis. Nano Lett. 2017;17(9):5193–5198. doi:10.1021/acs.nanolett.7b01006.61 Copyright (2017) American Chemical Society. (C) Self-assembly of aptamer-based nanohydrogel using Y-shaped building blocks for targeted cancer gene therapy. Adapted with permission from Li J, Zheng C, Cansiz S, et al. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J Am Chem Soc. 2015;137(4):1412–1415. doi:10.1021/ja512293f.54 Copyright ACS, https://pubs.acs.org/doi/abs/10.1021%2Fnn3022662. Further permissions related to the material excerpted should be directed to ACS. |
Hybrid hydrogels can lead to many preferred properties that would otherwise not be achievable by other nanostructures. These properties include biocompatibility, mass transport of ions, nutrients, proteins, drug, etc., biodegradability, mimicking dynamic nature of surrounding microenvironment (eg, extracellular matrix (ECM)), mechanical properties (porosity, stiffness, adhesion, swelling and phase transition (sol-gel) and stimuli-responsiveness.145 These smart polymers afford controlled disruption/deformation of the DNA network at the desired microenvironment to achieve on-target cargo release for a therapeutic effect (Figure 7A).146 From this point of view, tissue engineering and cell therapy for regenerative medicine can benefit from the soft nature of DNA hydrogels allowing potential imitation of the physiological environment and living matrices, while porous structure mainly benefits accommodation of therapeutic payloads.147
Different types of DNA building blocks can be used to produce these hydrogels, including single-stranded DNA branched double-stranded DNA, or Y-shaped DNA or X-shaped DNA through intermolecular i-motif structures, enzyme ligation, DNA hybridization, or enzyme polymerization. Although the use of DNA hydrogels in the clinic is not yet practiced, nonetheless, they already find numerous application in drug delivery146 sensing,148 tissue engineering,149,150 3D cell culture,149 providing a template for nanoparticle synthesis,92,151 and cell transplant therapy,152 among others (see for review).153
Hydrogels are popular nanostructures for tissue engineering because they can be tailor-made as biomimetics of ECM. DNA hydrogels are applicable for wound dressing with fluorescent and anti-bacterial potential. Due to their multifunctionality, they can simultaneously perform biosensing, and bioimaging tasks in living cells, as well.97,98,154
In terms of their biomedical applications, DNA gels nanostructures can be used for in situ encapsulation and preservation by protecting their cargoes from degradation, as reported for camptothecin (CPT), a DNA-binding drug, as well as porcine insulin.155 Interestingly, the formation of target-triggered DNA hydrogel has been harnessed as an effective tool for live single-cell isolation by liquid biopsy.61 For example, a DNA staple strand with aptamer-toehold biblocks is shown to specifically recognize EpCAM, a cell surface biomarker expressed by circulating tumor cells (CTCs). Binding of the aptamer to EpCAM triggers subsequent aptamer-triggered hybridization chain reaction (HCR) via toehold-initiated branch migration for simultaneous capturing and clocking of live single/clustered CTCs in porous DNA hydrogel with minimal cell damage. Moreover, controlled decloaking of CTCs was affordable using defined chemical stimuli to release intact living CTCs for subsequent culture and live cell analysis (Figure 7B).61
Interestingly, Dgel made of branched cross-linked DNA has found application for integration of plasmid-coding genes to envision a cell-free gene/protein synthesis system for production of different types of proteins/genes with extremely high production yields.156 In comparison to a linear plasmid, a high yield of over 300 times157 and ~50 times is reported for protein and mRNA expression, respectively.75 In view of the high RNA productivity of the Dgel, a recent study has reported a DNA hydrogel that can be used as RNA-producing machinery for gene regulation inside live cells. For this purpose, a plasmid carrying the gene transcribing siRNA against a target gene (green fluorescent protein (GFP)) was introduced into the gel scaffold. Such DNA hydrogel was capable to effectively shut down fluorescent protein expression in live cells.75
Another study, reported an albumin-DNA hybrid hydrogel for the controlled release of Rho-inhibiting C3 toxin for targeted inhibition of osteoclast formation and activity and possible application in local treatment of bone disease such as osteoporosis. This system is desirable with regard to its self-healing and injectable nature, DNA hybridization-mediated rapid gelation under physiological conditions, and the possibility to be loaded with specific cargoes (active proteins) and promote their spatiotemporal controlled release.67
With the aim of cancer-targeting, multifunctional and programmable aptamer-based DNA nanoassembly (AptNA) is reported using three Y-shaped building units; each incorporating different functional elements including targeting aptamer (sgc8), a therapeutic antisense oligonucleotide (MDR1 siRNA), and intercalated anticancer drug (Dox). This DNA nanohydrogel is a promising anti-cancer material due to its modularity, multifunctionality, programmability, excellent biocompatibility, and biostability. It also provides active targeting and controllable transportation of drug to the on-target cells. Moreover, using such a system drug resistance was reduced due to inhibition of P-gp expression –the anti-cancer drug efflux pump receptor–by the MDR1 siRNA.158 Similarly, catalytic hybridization was used for assembly of aptamer-based nanohydrogels (Y-gel-Apt), by employing two Y-shaped monomers (YMA, YMB) and a DNA linker (LK) bearing disulfide linkages and sticky ends. YMA contained a DNAzyme sequence for targeting MMP-9 and antisense oligonucleotide sequence in LK that enabled targeting of c-raf-1 –a tumor proliferation marker. Sticky ends were used for hybridization-mediated self-assembly whereas disulfide linkages led to the rapid fragmentation of nanohydrogels under cancer reductive environment, enabling a stimuli-responsive effective gene therapy (Figure 7C).54 More examples of aptamer-incorporated hydrogels for theranostics applications are overviewed elsewhere.
DNA Nanoswitch
Polymeric nanostructures such as hydrogels, liposomal/micellar nanocarriers or DNA-based hydrogels/nanostructures equipped with DNA switches such as azobenzene (Figure 8A)159/disulfide bonds are examples of static nanoswitches utilized for selective delivery of chemotherapeutics to cancer cells in response to environmental triggers such as hypoxia/pH/redox actuators.147 For example, disulfide-crosslinked nanohydrogels intercalated with Dox undergo fragmentation upon exposure to reducing agents dithiothreitol (DTT) or glutathione (GSH). These redox/pH dual stimuli-responsive nanohydrogels make promising candidates for delivering anti-cancer drugs because of their biocompatibility, adequate drug loading capacity, biodegradability, controlled yet fast drug release potential (Figure 8B).160 Moreover, bioreducible nanohydrogels-like nanocapsules (NCs) were reported to actively target cancer cells coated with folic acid.27
 |
Figure 8 DNA nanoswitch systems as cancer theranostics. (A) Azobenzene-integrated photo controlled drug release from DNA/mesoporous silica. Adapted with permission from YuanQ, ZhangY,Chen T, et al. Photon-manipulated drug release from a mesoporous nanocontainer controlled by azobenzene-modified nucleic acid. ACS Nano. 2012;6(7):6337–6344. doi:10.1021/nn3018365.159 Copyright (2012) American Chemical Society. (B) Redox stimuli-responsive drug release of the PMAA nanohydrogels. Adapted with permission from Pan YJ, Chen YY, Wang DR, et al. Redox/pH dual stimuli-responsive biodegradable nanohydrogels with varying responses to dithiothreitol and glutathione for controlled drug release. Biomaterials. 2012;33(27):6570–6579. doi:10.1016/j.biomaterials.2012.05.062, copyright (2012), with permission from Elsevier160 (C) Aptamer-based DNA nanoswitch for controlled drug release. Adapted with approval of Mo R, Jiang T, Sun W, Gu Z. ATP-responsive DNA-graphene hybrid nanoaggregates for anticancer drug delivery. Biomaterials. 2015;50:67–74. doi:10.1016/j.biomaterials.2015.01.053, copyright (2015) with permission from Elsevier.163 |
Light-switchable aptamer-based nanoconstructs were used for targeting and release of chemotherapeutics within the cancerous milieu. These complexes consisted of lipidated versions of a hepatocyte growth factor receptor (cMet) binding aptamer and a separate lipidated GC-rich DNA hairpin motif bearing intercalated Dox. Multiple 2’,6’-dimethylazobenzene moieties were incorporated into the Dox-binding motif to trigger the release of the chemotherapeutics by photoisomerization. The combined features of these nanocarriers increased serum nuclease resistance, favored their import into cells, allowed the selective photo-induced release of the chemotherapeutic into the targeted cells, and enhanced cell mortality.74 In addition, pH-responsive and light-responsive DNA microcapsules that encapsulated different loads including Dox-modified dextran, tetramethylrhodamine-modified dextran, CdSe/ZnS quantum dots, MP-11 and microperoxidase-11 has been described for controlled cargo release and cancer therapy in vitro.26 The pH-responsive microcapsules composed of a cytosine-rich layer cross-linked by nucleic acid bridges and the light-responsive microcapsules were made of photocleavable o-nitrobenzyl-phosphate-modified DNA shells.26
In another study, reduction-responsive hydrogel nanoparticles are utilized for siRNA delivery and targeted gene silencing.53 Furthermore, near-infrared light-responsive hybrid drug delivery platform composed of Au–Ag nanorods (Au–Ag NRs) and DNA cross-linked polymeric shell was reported in which near infra-red (NIR) irradiation induce photothermal effect of the Au–Ag NRs which leads to rapid heating of the nanogel, and promote programmed and swift release of captured drugs into the target environment.161 Likewise, reversible DNA hybridization allows specific DNA oligonucleotides to act as triggers for autonomous switching of nanostructures to load or release the therapeutic targets. For example, aptamer-cross linked hydrogels were utilized to control the release of drugs and nanoparticles in response to binding of the DNA aptamer to small metabolites such as adenosine, AMP, and ATP (Figure 8C).162,163
Conclusion And Outlook
Since its inception in the early 1980s and the invention of DNA origami in 2006, DNA nanotechnology has made remarkable strides towards a veriety of applications. In view of our interest in the era of cancer research, we provided an overview of the advances in DNA nanotechnology at the interface of static DNA-based devices and their broad applications in the field of bio/nanomedicine (Table 1). Compared to other nanomaterials, such as polymeric, organic, metallic, and lipid-based nanoparticles, etc., the beauty of DN can be attributed to the simple principle of base pairing that allows construction of an unlimited number of nanostructures with different sizes, shapes, charges, and modularity to serve for a specific purpose. For example, DNA polyhedra (DNA box), and hydrogel are both porous structures that can accommodate therapeutic agent inside while functional ligands/moieties can be coated on the surface of DN. DNA box toolkits are meant for on-command opening (provided by aptamer) and exposure of cloaked cytotoxic drugs, when only the correct trigger (eg, ATP) is available, allowing effective on-target drug delivery. In addition to its function as a drug carrier, the hydrogel structure offers an additional advantage as it can undergo reversible changes of shape (gel/sol state), size (shrunken/swollen) and charge (deprotonated/protonated). Such potential to capture and adapt to changes in the microenvironment is important for the design of nanocarriers taking into account the dynamic and heterogeneous microenvironment of cancer cells. For example, pH-responsive hydrogel (provided by azo-linker) allows the shrunk hydrogel to enter the endo-lysosomes where the size expansion occurs within the low pH endo-lysosomal vesicles, resulting in a larger pore size and thus the leakage of cytotoxic drugs to induce cancer cell apoptosis. Meanwhile, acid-triggered protonated hydrogel that results in size expansion and electrostatic repulsion of DN promotes endo-lysosomal burst release of hydrogel into the cytosol. Under normal pH, the hydrogel can shrink back to its original size, and its subsequent transport outside the dead cell starts another cycle of penetration into the nearby cancer cells.164 This way, recycling hydrogel can penetrate into the deeper regions of the tumor by peeling off tumor layers one after another to envision an enhanced therapeutic index for cancer therapy.
With regard to the nature of DNA, the advantages of DNA based architectures including a rapid and facile self-assembly method, forming rigid yet complex structures, automation, and self-amplification potential makes it a suitable and addressable material with unlimited applications in biology, medicine and especially preclinical cancer research and therapy. However, for DNA nanotechnology to reach its full potential, several limitations should be addressed, including simplified and automated design platforms, straightforward and economical production methods, scale-up strategies for size expansion, efficient chemical functionality, enhanced stability to survive harsh fabrication environments, minimizing immunogenicity and assembly defects, and finally improving their penetration and cellular uptake through physiological barriers.
Together, the facile assembly of DNA structures to well-defined architectures capable of crossing physiological barriers, being stable in biological fluids inside the body plus potential for biosensing and performing tasks on target cells with nanometer precision, if achieved, can deliver an unprecedented opportunity for timely cancer detection and therapy at the level of one single cell.
Acknowledgments
The authors would like to sincerely thank Dr. Thorsten Lars Schmidt for his valuable comments and fruitful discussions. The APC for this research was funded by Dr. Peyman Zare from the Faculty of Medicine, Cardinal Stefan Wyszyński University in Warsaw, 01-938 Warsaw, Poland. Tahereh Javaheri no longer works at Ludwig Boltzmann Institute.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jahanban-Esfahlan R, Seidi K, Banimohamad-Shotorbani B, Jahanban-Esfahlan A, Yousefi B. Combination of nanotechnology with vascular targeting agents for effective cancer therapy. J Cell Physiol. 2017;233(4):2982–2992. doi:10.1002/jcp.26051
2. Seidi K, Neubauer HA, Moriggl R, Jahanban-Esfahlan R, Javaheri T. Tumor target amplification: implications for nano drug delivery systems. J Control Release. 2018;275:142–161. doi:10.1016/j.jconrel.2018.02.020
3. Jahanban-Esfahlan A, Ostadrahimi A, Jahanban-Esfahlan R, Roufegarinejad L, Tabibiazar M, Amarowicz R. Recent developments in the detection of bovine serum albumin. Int J Biol Macromol. 2019;138:602–617. doi:10.1016/j.ijbiomac.2019.07.096
4. Dianat-Moghadam H, Heydarifard M, Jahanban-Esfahlan R, et al. Cancer stem cells-emanated therapy resistance: implications for liposomal drug delivery systems. J Control Release. 2018;288:62–83. doi:10.1016/j.jconrel.2018.08.043
5. Jahanban-Esfahlan R, de la Guardia M, Ahmadi D, Yousefi B. Modulating tumor hypoxia by nanomedicine for effective cancer therapy. J Cell Physiol. 2017;233(3):2019–2031. doi:10.1002/jcp.25859
6. Seeman NC. DNA in a material world. Nature. 2003;421:427. doi:10.1038/nature01406
7. Goodman RP, Berry RM, Turberfield AJ. The single-step synthesis of a DNA tetrahedron. Chem Commun (Camb). 2004;12:1372–1373. doi:10.1039/b402293a
8. Herrera VLM, Colby AH, Ruiz-Opazo N, Coleman DG, Grinstaff MW. Nucleic acid nanomedicines in Phase II/III clinical trials: translation of nucleic acid therapies for reprogramming cells. Nanomedicine. 2018;13(16):2083–2098. doi:10.2217/nnm-2018-0122
9. Bhatia D, Surana S, Chakraborty S, Koushika SP, Krishnan Y. A synthetic icosahedral DNA-based host–cargo complex for functional in vivo imaging. Nat Commun. 2011;2:339. doi:10.1038/ncomms1337
10. Hu R, Zhang X, Zhao Z, et al. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew Chem Int Ed Engl. 2014;53(23):5821–5826. doi:10.1002/anie.201400323
11. Jiang D, Ge Z, Im H-J, et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat Biomed Eng. 2018;2(11):865–877. doi:10.1038/s41551-018-0317-8
12. Wang M. DNA origami scavenges ROS in the kidney. Nature Reviews Nephrology. 2019;15(61).
13. Zhang L, Jean SR, Ahmed S, et al. Multifunctional quantum dot DNA hydrogels. Nat Commun. 2017;8(1):381. doi:10.1038/s41467-017-00298-w
14. Jiang D, Sun Y, Li J, et al. Multiple-armed tetrahedral DNA nanostructures for tumor-targeting, dual-modality in vivo imaging. ACS Appl Mater Interfaces. 2016;8(7):4378–4384. doi:10.1021/acsami.5b10792
15. Li L, Niu C, Li T, et al. Ultrasensitive electrochemiluminescence biosensor for detection of laminin based on DNA dendrimer-carried luminophore and DNA nanomachine-mediated target recycling amplification. Biosens Bioelectron. 2018;101:206–212. doi:10.1016/j.bios.2017.10.009
16. Wang K, You M, Chen Y, et al. Self-assembly of a bifunctional DNA carrier for drug delivery. Angew Chem Int Ed Engl. 2011;50(27):6098–6101. doi:10.1002/anie.201008053
17. Shieh YA, Yang SJ, Wei MF, Shieh MJ. Aptamer-based tumor-targeted drug delivery for photodynamic therapy. ACS Nano. 2010;4(3):1433–1442. doi:10.1021/nn901374b
18. Liu T, Wang C, Gu X, et al. Drug delivery with PEGylated MoS2 nano-sheets for combined photothermal and chemotherapy of cancer. Adv Mater. 2014;26(21):3433–3440. doi:10.1002/adma.201305256
19. Jiang Q, Song C, Nangreave J, et al. DNA origami as a carrier for circumvention of drug resistance. J Am Chem Soc. 2012;134(32):13396–13403. doi:10.1021/ja304263n
20. Zhang C, Li X, Tian C, et al. DNA nanocages swallow gold nanoparticles (AuNPs) to form AuNP@DNA cage core-shell structures. ACS Nano. 2014;8(2):1130–1135. doi:10.1021/nn406039p
21. Zhao YX, Shaw A, Zeng X, Benson E, Nystrom AM, Hogberg B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano. 2012;6(10):8684–8691. doi:10.1021/nn3022662
22. Sun W, Jiang T, Lu Y, Reiff M, Mo R, Gu Z. Cocoon-like self-degradable DNA nanoclew for anticancer drug delivery. J Am Chem Soc. 2014;136(42):14722–14725. doi:10.1021/ja5088024
23. Kumar V, Bayda S, Hadla M, et al. Enhanced chemotherapeutic behavior of open-caged DNA@doxorubicin nanostructures for cancer cells. J Cell Physiol. 2016;231(1):106–110. doi:10.1002/jcp.25057
24. Song J, Im K, Hwang S, et al. DNA hydrogel delivery vehicle for light-triggered and synergistic cancer therapy. Nanoscale. 2015;7(21):9433–9437. doi:10.1039/c5nr00858a
25. Song J, Hwang S, Im K, et al. Light-responsible DNA hydrogel–gold nanoparticle assembly for synergistic cancer therapy. J Mater Chem B. 2015;3(8):1537–1543. doi:10.1039/C4TB01519C
26. Huang F, Liao WC, Sohn YS, Nechushtai R, Lu CH, Willner I. Light-responsive and pH-responsive DNA microcapsules for controlled release of loads. J Am Chem Soc. 2016;138(28):8936–8945. doi:10.1021/jacs.6b04773
27. Yi Q, Ma J, Kang K, Gu Z. Bioreducible nanocapsules for folic acid-assisted targeting and effective tumor-specific chemotherapy. Int J Nanomedicine. 2018;13:653–667. doi:10.2147/IJN.S149458
28. Taghdisi SM, Danesh NM, Ramezani M, et al. Double targeting and aptamer-assisted controlled release delivery of epirubicin to cancer cells by aptamers-based dendrimer in vitro and in vivo. Eur J Pharm Biopharm. 2016;102:152–158. doi:10.1016/j.ejpb.2016.03.013
29. Wang YM, Wu Z, Liu SJ, Chu X. Structure-switching aptamer triggering hybridization chain reaction on the cell surface for activatable theranostics. Anal Chem. 2015;87(13):6470–6474. doi:10.1021/acs.analchem.5b01634
30. Halley PD, Lucas CR, McWilliams EM, et al. Daunorubicin-loaded DNA origami nanostructures circumvent drug-resistance mechanisms in a leukemia model. Small. 2016;12(3):308–320. doi:10.1002/smll.201502118
31. Setyawati MI, Kutty RV, Tay CY, Yuan X, Xie J, Leong DT. Novel theranostic DNA nanoscaffolds for the simultaneous detection and killing of escherichia coli and staphylococcus aureus. ACS Appl Mater Interfaces. 2014;6(24):21822–21831. doi:10.1021/am502591c
32. Ren J, Hu Y, Lu CH, et al. pH-responsive and switchable triplex-based DNA hydrogels. Chem Sci. 2015;6(7):4190–4195. doi:10.1039/c5sc00594a
33. Schüller VJ, Heidegger S, Sandholzer N, et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano. 2011;5(12):9696–9702. doi:10.1021/nn203161y
34. Qu Y, Yang J, Zhan P, et al. Self-assembled DNA dendrimer nanoparticle for efficient delivery of immunostimulatory CpG motifs. ACS Appl Mater Interfaces. 2017;9(24):20324–20329. doi:10.1021/acsami.7b05890
35. Li J, Pei H, Zhu B, et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5(11):8783–8789. doi:10.1021/nn202774x
36. Zhang L, Zhu G, Mei L, et al. Self-assembled DNA immunonanoflowers as multivalent CpG nanoagents. ACS Appl Mater Interfaces. 2015;7(43):24069–24074. doi:10.1021/acsami.5b06987
37. Chen N, Wei M, Sun Y, et al. Self-assembly of poly-adenine-tailed CpG oligonucleotide-gold nanoparticle nanoconjugates with immunostimulatory activity. Small. 2014;10(2):368–375. doi:10.1002/smll.201300903
38. Wang C, Sun W, Wright G, Wang AZ, Gu Z. Inflammation-triggered cancer immunotherapy by programmed delivery of CpG and anti-PD1 antibody. Adv Mater. 2016;28(40):8912–8920. doi:10.1002/adma.201506312
39. Sellner S, Kocabey S, Nekolla K, Krombach F, Liedl T, Rehberg M. DNA nanotubes as intracellular delivery vehicles in vivo. Biomaterials. 2015;53:453–463. doi:10.1016/j.biomaterials.2015.02.099
40. Ohtsuki S, Matsuzaki N, Mohri K, et al. Optimal arrangement of four short DNA strands for delivery of immunostimulatory nucleic acids to immune cells. Nucleic Acid Ther. 2015;25(5):245–253. doi:10.1089/nat.2014.0524
41. Nishikawa M, Mizuno Y, Mohri K, et al. Biodegradable CpG DNA hydrogels for sustained delivery of doxorubicin and immunostimulatory signals in tumor-bearing mice. Biomaterials. 2011;32(2):488–494. doi:10.1016/j.biomaterials.2010.09.013
42. Zhang Y, Ma W, Zhu Y, et al. Inhibiting methicillin-resistant staphylococcus aureus by tetrahedral DNA nanostructure-enabled antisense peptide nucleic acid delivery. Nano Lett. 2018;18:5652–5659.
43. Readman JB, Dickson G, Coldham NG. Tetrahedral DNA nanoparticle vector for intracellular delivery of targeted peptide nucleic acid antisense agents to restore antibiotic sensitivity in cefotaxime-resistant escherichia coli. Nucleic Acid Ther. 2017;27(3):176–181. doi:10.1089/nat.2016.0644
44. Qian H, Tay CY, Setyawati MI, Chia SL, Lee DS, Leong DT. Protecting microRNAs from RNase degradation with steric DNA nanostructures. Chem Sci. 2017;8(2):1062–1067. doi:10.1039/c6sc01829g
45. Liu Q, Wang D, Yuan M, et al. Capturing intracellular oncogenic microRNAs with self-assembled DNA nanostructures for microRNA-based cancer therapy. Chem Sci. 2018;9(38):7562–7568. doi:10.1039/c8sc03039a
46. Nahar S, Nayak AK, Ghosh A, Subudhi U, Maiti S. Enhanced and synergistic downregulation of oncogenic miRNAs by self-assembled branched DNA. Nanoscale. 2017;10(1):195–202. doi:10.1039/c7nr06601e
47. Shin SW, Lee BS, Yang K, et al. Fluorescence-coded DNA nanostructure probe system to enable discrimination of tumor heterogeneity via a screening of dual intracellular microRNA signatures in situ. Sci Rep. 2017;7(1):13499. doi:10.1038/s41598-017-13456-3
48. Chen G, Liu D, He C, Gannett TR, Lin W, Weizmann Y. Enzymatic synthesis of periodic DNA nanoribbons for intracellular pH sensing and gene silencing. J Am Chem Soc. 2015;137(11):3844–3851. doi:10.1021/ja512665z
49. Lee H, Lytton-Jean AK, Chen Y, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7(6):389–393. doi:10.1038/nnano.2012.73
50. Bujold KE, Hsu JCC, Sleiman HF, Optimized DNA. “Nanosuitcases” for encapsulation and conditional release of siRNA. J Am Chem Soc. 2016;138(42):14030–14038. doi:10.1021/jacs.6b08369
51. Esteban-Fernández de Ávila B, Angell C, Soto F, et al. Acoustically propelled nanomotors for intracellular siRNA delivery. ACS Nano. 2016;10(5):4997–5005. doi:10.1021/acsnano.6b01415
52. Hong CA, Eltoukhy AA, Lee H, Langer R, Anderson DG, Nam YS. Dendrimeric siRNA for efficient gene silencing. Angew Chem Int Ed Engl. 2015;54(23):6740–6744. doi:10.1002/anie.201412493
53. Dunn SS, Tian S, Blake S, et al. Reductively responsive siRNA-conjugated hydrogel nanoparticles for gene silencing. J Am Chem Soc. 2012;134(17):7423–7430. doi:10.1021/ja300174v
54. Li J, Zheng C, Cansiz S, et al. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J Am Chem Soc. 2015;137(4):1412–1415. doi:10.1021/ja512293f
55. Fakhoury JJ, McLaughlin CK, Edwardson TW, Conway JW, Sleiman HF. Development and characterization of gene silencing DNA cages. Biomacromolecules. 2014;15(1):276–282. doi:10.1021/bm401532n
56. Sun W, Ji W, Hall JM, et al. Efficient delivery of CRISPR-Cas9 for genome editing via self-assembled DNA nanoclews. Angew Chem Int Ed Engl. 2015;54(41):12029–12033. doi:10.1002/anie.201506030
57. Charoenphol P, Bermudez H. Aptamer-targeted DNA nanostructures for therapeutic delivery. Mol Pharm. 2014;11(5):1721–1725. doi:10.1021/mp500047b
58. Chang M, Yang C-S, Huang D-M. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano. 2011;5(8):6156–6163. doi:10.1021/nn200693a
59. Walsh AS, Yin H, Erben CM, Wood MJ, Turberfield AJ. DNA cage delivery to mammalian cells. ACS Nano. 2011;5(7):5427–5432. doi:10.1021/nn2005574
60. Meng HM, Zhang X, Lv Y, et al. DNA dendrimer: an efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano. 2014;8(6):6171–6181. doi:10.1021/nn5015962
61. Song P, Ye D, Zuo X, et al. DNA hydrogel with aptamer-toehold-based recognition, cloaking, and decloaking of circulating tumor cells for live cell analysis. Nano Lett. 2017;17(9):5193–5198. doi:10.1021/acs.nanolett.7b01006
62. Ma Y, Mao Y, An Y, et al. Target-responsive DNA hydrogel for non-enzymatic and visual detection of glucose. Analyst. 2018;143(7):1679–1684. doi:10.1039/c8an00010g
63. Ji Y, Zhang L, Zhu L, Lei J, Wu J, Ju H. Binding-induced DNA walker for signal amplification in highly selective electrochemical detection of protein. Biosens Bioelectron. 2017;96:201–205. doi:10.1016/j.bios.2017.05.008
64. Li F, Cha TG, Pan J, Ozcelikkale A, Han B, Choi JH. DNA walker-regulated cancer cell growth inhibition. Chembiochem. 2016;17(12):1138–1141. doi:10.1002/cbic.201600052
65. Wang S, Ji Y, Fu H, Ju H, Lei J. A rolling circle amplification-assisted DNA walker triggered by multiple DNAzyme cores for highly sensitive electrochemical biosensing. Analyst. 2019;144:691–697. doi:10.1039/c8an01892h
66. Wang K, He M-Q, Zhai F-H, Wang J, He R-H, Yu Y-L. Autonomous DNA nanomachine based on cascade amplification of strand displacement and DNA walker for detection of multiple DNAs. Biosens Bioelectron. 2018;105:159–165. doi:10.1016/j.bios.2018.01.044
67. Gacanin J, Kovtun A, Fischer S, et al. Spatiotemporally controlled release of rho-inhibiting C3 toxin from a protein-DNA hybrid hydrogel for targeted inhibition of osteoclast formation and activity. Adv Healthc Mater. 2017;6(21). doi:10.1002/adhm.201700392.
68. Zhu G, Mei L, Vishwasrao HD, et al. Intertwining DNA-RNA nanocapsules loaded with tumor neoantigens as synergistic nanovaccines for cancer immunotherapy. Nat Commun. 2017;8(1):1482. doi:10.1038/s41467-017-01386-7
69. Zheng J, Li N, Li C, et al. A nonenzymatic DNA nanomachine for biomolecular detection by target recycling of hairpin DNA cascade amplification. Biosens Bioelectron. 2018;107:40–46. doi:10.1016/j.bios.2018.01.054
70. Feng Q, Zhao X, Guo Y, Liu M, Wang P. Stochastic DNA walker for electrochemical biosensing sensitized with gold nanocages@graphene nanoribbons. Biosens Bioelectron. 2018;108:97–102. doi:10.1016/j.bios.2018.02.050
71. Zhang H, Chao J, Pan D, et al. DNA origami-based shape IDs for single-molecule nanomechanical genotyping. Nat Commun. 2017;8:14738. doi:10.1038/ncomms14738
72. Liu K, Pan D, Wen Y, et al. Identifying the genotypes of hepatitis B virus (HBV) with DNA origami label. Small. 2018;14:6.
73. Na W, Nam D, Lee H, Shin S. Rapid molecular diagnosis of infectious viruses in microfluidics using DNA hydrogel formation. Biosens Bioelectron. 2018;108:9–13. doi:10.1016/j.bios.2018.02.040
74. Prusty DK, Adam V, Zadegan RM, Irsen S, Famulok M. Supramolecular aptamer nano-constructs for receptor-mediated targeting and light-triggered release of chemotherapeutics into cancer cells. Nat Commun. 2018;9(1):535. doi:10.1038/s41467-018-02929-2
75. Song J, Lee M, Kim T, et al. A RNA producing DNA hydrogel as a platform for a high performance RNA interference system. Nat Commun. 2018;9(1):4331. doi:10.1038/s41467-018-06864-0
76. Peng H, Li XF. A microRNA-initiated DNAzyme motor operating in living cells. Nat Commun. 2017;8:14378. doi:10.1038/ncomms14378
77. Hwang MT, Wang Z, Ping J, et al. DNA nanotweezers and graphene transistor enable label-free genotyping. Adv Mater. 2018;30:e1802440. doi:10.1002/adma.v30.34
78. Bath J, Green SJ, Turberfield AJ. A free-running DNA motor powered by a nicking enzyme. Angew Chem Int Ed Engl. 2005;44(28):4358–4361. doi:10.1002/anie.200501262
79. Xu X, Wang L, Li K, Huang Q, Jiang W. A smart DNA tweezer for detection of human telomerase activity. Anal Chem. 2018;90(5):3521–3530. doi:10.1021/acs.analchem.7b05373
80. Lilienthal S, Shpilt Z, Wang F, Orbach R, Willner I. Programmed DNAzyme-triggered dissolution of DNA-based hydrogels: means for controlled release of biocatalysts and for the activation of enzyme cascades. ACS Appl Mater Interfaces. 2015;7(16):8923–8931. doi:10.1021/acsami.5b02156
81. Yata T, Takahashi Y, Tan M, et al. DNA nanotechnology-based composite-type gold nanoparticle-immunostimulatory DNA hydrogel for tumor photothermal immunotherapy. Biomaterials. 2017;146:136–145. doi:10.1016/j.biomaterials.2017.09.014
82. Elbaz J, Wang Z-G, Orbach R, Willner I. pH-stimulated concurrent mechanical activation of two DNA “Tweezers”. A “SET−RESET” logic gate system. Nano Lett. 2009;9(12):4510–4514. doi:10.1021/nl902859m
83. Crawford R, Erben CM, Periz J, et al. Non-covalent single transcription factor encapsulation inside a DNA cage. Angew Chem Int Ed Engl. 2013;52(8):2284–2288. doi:10.1002/anie.201207914
84. Porchetta A, Ippodrino R, Marini B, Caruso A, Caccuri F, Ricci F. Programmable nucleic acid nanoswitches for the rapid, single-step detection of antibodies in bodily fluids. J Am Chem Soc. 2018;140(3):947–953. doi:10.1021/jacs.7b09347
85. Burns JR, Lamarre B, Pyne ALB, Noble JE, Ryadnov MG. DNA origami inside-out viruses. ACS Synth Biol. 2018;7(3):767–773. doi:10.1021/acssynbio.7b00278
86. Stephanopoulos N, Freeman R, North HA, et al. Bioactive DNA-peptide nanotubes enhance the differentiation of neural stem cells into neurons. Nano Lett. 2015;15(1):603–609. doi:10.1021/nl504079q
87. Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335(6070):831–834. doi:10.1126/science.1214081
88. Wang Y, Jiang L-P, Zhou S, Bi S, Zhu -J-J. DNA polymerase-directed hairpin assembly for targeted drug delivery and amplified biosensing. ACS Appl Mater Interfaces. 2016;8(40):26532–26540. doi:10.1021/acsami.6b08597
89. Zhao Z, Fu J, Dhakal S, et al. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat Commun. 2016;7:10619. doi:10.1038/ncomms10619
90. Ora A, Järvihaavisto E, Zhang H, et al. Cellular delivery of enzyme-loaded DNA origami. Chem Commun. 2016;52(98):14161–14164. doi:10.1039/c6cc08197e
91. Liu M, Fu J, Hejesen C, et al. A DNA tweezer-actuated enzyme nanoreactor. Nat Commun. 2013;4:2127. doi:10.1038/ncomms3127
92. Hur J, Im K, Kim SW, et al. DNA hydrogel templated carbon nanotube and polyaniline assembly and its applications for electrochemical energy storage devices. J Mater Chem A. 2013;1(46):14460–14466. doi:10.1039/c3ta13382f
93. Zhang Z-M, Gao P-C, Wang Z-F, Sun B-W, Jiang Y. DNA-caged gold nanoparticles for controlled release of doxorubicin triggered by a DNA enzyme and pH. Chem Commun. 2015;51(65):12996–12999. doi:10.1039/c5cc05164a
94. Zhu D, Liu W, Cao W, et al. Multiple amplified electrochemical detection of microRNA-21 using hierarchical flower-like gold nanostructures combined with gold-enriched hybridization chain reaction. Electroanalysis. 2018;30(7):1349–1356. doi:10.1002/elan.201700696
95. Dong S, Zhao R, Zhu J, et al. Electrochemical DNA biosensor based on a tetrahedral nanostructure probe for the detection of avian influenza A (H7N9) virus. ACS Appl Mater Interfaces. 2015;7(16):8834–8842. doi:10.1021/acsami.5b01438
96. Arnon S, Dahan N, Koren A, et al. Thought-controlled nanoscale robots in a living host. PLoS One. 2016;11(8):e0161227. doi:10.1371/journal.pone.0161227
97. Geng J, Yao C, Kou X, Tang J, Luo D, Yang D. A fluorescent biofunctional DNA hydrogel prepared by enzymatic polymerization. Adv Healthc Mater. 2018;7(5):1700998. doi:10.1002/adhm.v7.5
98. Li J, Yu J, Huang Y, Zhao H, Tian L. Highly stable and multiemissive silver nanoclusters synthesized in situ in a DNA hydrogel and their application for hydroxyl radical sensing. ACS Appl Mater Interfaces. 2018;10(31):26075–26083. doi:10.1021/acsami.8b09152
99. Udomprasert A, Kangsamaksin T. DNA origami applications in cancer therapy. Cancer Sci. 2017;108(8):1535–1543. doi:10.1111/cas.13290
100. Perrault SD, Shih WM. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano. 2014;8(5):5132–5140. doi:10.1021/nn5011914
101. Mei Q, Wei X, Su F, et al. Stability of DNA origami nanoarrays in cell lysate. Nano Lett. 2011;11(4):1477–1482. doi:10.1021/nl1040836
102. Meng M, Schmidtgall B, Ducho C. Enhanced stability of DNA oligonucleotides with partially zwitterionic backbone structures in biological media. Molecules. 2018;23(11):2941. doi:10.3390/molecules23112941
103. Zhuang X, Ma X, Xue X, et al. A photosensitizer-loaded DNA origami nanosystem for photodynamic therapy. ACS Nano. 2016;10(3):3486–3495. doi:10.1021/acsnano.5b07671
104. Schmidtgall B, Kuepper A, Meng M, Grossmann TN, Ducho C. Oligonucleotides with cationic backbone and their hybridization with DNA: interplay of base pairing and electrostatic attraction. Chemistry.2018;24(7):1544–1553. doi:10.1002/chem.201704338
105. Zadegan RM, Jepsen MDE, Hildebrandt LL, Birkedal V, Kjems J. Construction of a fuzzy and boolean logic gates based on DNA. Small. 2015;11(15):1811–1817. doi:10.1002/smll.201402755
106. Green CM, Schutt K, Morris N, et al. Metrology of DNA arrays by super-resolution microscopy. Nanoscale. 2017;9(29):10205–10211. doi:10.1039/c7nr00928c
107. Zadegan RM, Lindau EG, Klein WP, et al. Twisting of DNA Origami from Intercalators. Sci Rep. 2017;7(1):7382. doi:10.1038/s41598-017-07796-3
108. Zhang Q, Jiang Q, Li N, et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano. 2014;8(7):6633–6643. doi:10.1021/nn502058j
109. Tinnefeld P, Acuna GP, Wei Q, et al. DNA origami nanotools for single-molecule biosensing and superresolution microscopy.
110. Shaw A, Hoffecker IT, Smyrlaki I, et al. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat Nanotechnol. 2019;14(2):184–190. doi:10.1038/s41565-018-0336-3
111. Hawkes W, Huang D, Reynolds P, et al. Probing the nanoscale organisation and multivalency of cell surface receptors: DNA origami nanoarrays for cellular studies with single-molecule control. Faraday Discuss. 2019. doi:10.1039/C9FD00023B
112. Huang D, Patel K, Perez-Garrido S, Marshall JF, Palma M. DNA origami nanoarrays for multivalent investigations of cancer cell spreading with nanoscale spatial resolution and single-molecule control. ACS Nano. 2019;13(1):728–736. doi:10.1021/acsnano.8b08010
113. Lu X, Liu J, Wu X, Ding B. Multifunctional DNA origami nanoplatforms for drug delivery. Chem Asian J. 2019;14(13):2193–2202. doi:10.1002/asia.201900574
114. Goodman RP, Schaap IA, Tardin CF, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310(5754):1661–1665. doi:10.1126/science.1120367
115. Karimi M, Zangabad PS, Mehdizadeh F, et al. Nanocaged platforms: modification, drug delivery and nanotoxicity. Opening synthetic cages to release the tiger. Nanoscale. 2017;9(4):1356–1392. doi:10.1039/c6nr07315h
116. Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew Chem Int Ed Engl. 2006;45(44):7414–7417. doi:10.1002/anie.200603392
117. Brodin JD, Sprangers AJ, McMillan JR, Mirkin CA. DNA-mediated cellular delivery of functional enzymes. J Am Chem Soc. 2015;137(47):14838–14841. doi:10.1021/jacs.5b09711
118. Kim KR, Kim DR, Lee T, et al. Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells. Chem Commun (Camb). 2013;49(20):2010–2012. doi:10.1039/c3cc38693g
119. Vindigni G, Raniolo S, Ottaviani A, et al. Receptor-mediated entry of pristine octahedral DNA nanocages in mammalian cells. ACS Nano. 2016;10(6):5971–5979. doi:10.1021/acsnano.6b01402
120. Raniolo S, Vindigni G, Ottaviani A, et al. Selective targeting and degradation of doxorubicin-loaded folate-functionalized DNA nanocages. Nanomedicine. 2018;14:1181–1190. doi:10.1016/j.nano.2018.02.002
121. Kim KR, Kim HY, Lee YD, et al. Self-assembled mirror DNA nanostructures for tumor-specific delivery of anticancer drugs. J Control Release. 2016;243:121–131. doi:10.1016/j.jconrel.2016.10.015
122. Xia Z, Wang P, Liu X, et al. Tumor-penetrating peptide-modified DNA tetrahedron for targeting drug delivery. Biochemistry. 2016;55(9):1326–1331. doi:10.1021/acs.biochem.5b01181
123. Goodman RP, Heilemann M, Doose S, Erben CM, Kapanidis AN, Turberfield AJ. Reconfigurable, braced, three-dimensional DNA nanostructures. Nat Nanotechnol. 2008;3(2):93–96. doi:10.1038/nnano.2008.3
124. Lee H, Lytton-Jean AKR, Chen Y, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7:389. doi:10.1038/nnano.2012.73
125. Vellampatti S, Heo R, Mitta Sekhar B, Park Jae H, Park Sung H. Aptamer-conjugated DNA nano-ring as the carrier of drug molecules. Nanotechnology. 2018;29(9):095602. doi:10.1088/1361-6528/aaa3cb
126. Pei H, Lu N, Wen Y, et al. A DNA nanostructure-based biomolecular probe carrier platform for electrochemical biosensing. Adv Mater. 2010;22(42):4754–4758. doi:10.1002/adma.201002767
127. Fu Y, Zeng D, Chao J, et al. Single-step rapid assembly of DNA origami nanostructures for addressable nanoscale bioreactors. J Am Chem Soc. 2013;135(2):696–702. doi:10.1021/ja3076692
128. Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 2006;6(7):1502–1504. doi:10.1021/nl060994c
129. Lin C, Ke Y, Liu Y, Mertig M, Gu J, Yan H. Functional DNA nanotube arrays: bottom-up meets top-down. Angew Chem Int Ed Engl. 2007;46(32):6089–6092. doi:10.1002/anie.200701767
130. Sacca B, Meyer R, Erkelenz M, et al. Orthogonal protein decoration of DNA origami. Angew Chem Int Ed Engl. 2010;49(49):9378–9383. doi:10.1002/anie.201005931
131. Nakata E, Liew FF, Uwatoko C, et al. Zinc-finger proteins for site-specific protein positioning on DNA-origami structures. Angew Chem Int Ed Engl. 2012;51(10):2421–2424. doi:10.1002/anie.201108199
132. Pinto YY, Le JD, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Sequence-encoded self-assembly of multiple-nanocomponent arrays by 2D DNA scaffolding. Nano Lett. 2005;5(12):2399–2402. doi:10.1021/nl0515495
133. Kuzuya A, Kimura M, Numajiri K, et al. Precisely programmed and robust 2D streptavidin nanoarrays by using periodical nanometer-scale wells embedded in DNA origami assembly. Chembiochem. 2009;10(11):1811–1815. doi:10.1002/cbic.200900229
134. Baker YR, Chen J, Brown J, et al. Preparation and characterization of manganese, cobalt and zinc DNA nanoflowers with tuneable morphology, DNA content and size. Nucleic Acids Res. 2018;46(15):7495–7505. doi:10.1093/nar/gky630
135. Shi L, Mu C, Gao T, et al. DNA nanoflower blooms in nanochannels: a new strategy for miRNA detection. Chem Commun. 2018;54(81):11391–11394. doi:10.1039/c8cc05690k
136. Lv Y, Hu R, Zhu G, et al. Preparation and biomedical applications of programmable and multifunctional DNA nanoflowers. Nat Protoc. 2015;10(10):1508–1524. doi:10.1038/nprot.2015.078
137. Mei L, Zhu G, Qiu L, et al. Self-assembled multifunctional DNA nanoflowers for the circumvention of multidrug resistance in targeted anticancer drug delivery. Nano Res. 2015;8(11):3447–3460.
138. Altinkaynak C, Tavlasoglu S, Ozdemir N, Ocsoy I. A new generation approach in enzyme immobilization: organic-inorganic hybrid nanoflowers with enhanced catalytic activity and stability. Enzyme Microb Technol. 2016;93–94:105–112. doi:10.1016/j.enzmictec.2016.06.011
139. Ge J, Lei J, Zare RN. Protein-inorganic hybrid nanoflowers. Nat Nanotechnol. 2012;7(7):428–432. doi:10.1038/nnano.2012.80
140. Zhao Y, Hu S, Wang H, et al. DNA dendrimer–streptavidin nanocomplex: an efficient signal amplifier for construction of biosensing platforms. Anal Chem. 2017;89(12):6907–6914. doi:10.1021/acs.analchem.7b01551
141. Lv Y, Peng R, Zhou Y, Zhang X, Tan W. Catalytic self-assembly of a DNA dendritic complex for efficient gene silencing. Chem Commun (Camb). 2016;52(7):1413–1415. doi:10.1039/c5cc06937h
142. Mohri K, Kusuki E, Ohtsuki S, et al. Self-assembling DNA dendrimer for effective delivery of immunostimulatory CpG DNA to immune cells. Biomacromolecules. 2015;16(4):1095–1101. doi:10.1021/bm501731f
143. Liu H, Cao T, Xu Y, Dong Y, Liu D. Tuning the mechanical properties of a DNA hydrogel in three phases based on ATP aptamer. Int J Mol Sci. 2018;19(6):1633.
144. Shimomura S, Nishimura T, Ogura Y, Tanida J. Photothermal fabrication of microscale patterned DNA hydrogels. R Soc Open Sci. 2018;5(2):171779. doi:10.1098/rsos.172483
145. Radvar E, Azevedo HS. Supramolecular peptide/polymer hybrid hydrogels for biomedical applications. Macromol Biosci. 2018;19:e1800221.
146. Kang H, Liu H, Zhang X, et al. Photoresponsive DNA-cross-linked hydrogels for controllable release and cancer therapy. Langmuir. 2011;27(1):399–408. doi:10.1021/la1037553
147. Gupta P, Vermani K, Garg S. Hydrogels: from controlled release to pH-responsive drug delivery. Drug Discov Today. 2002;7(10):569–579.
148. Zhu Z, Wu C, Liu H, et al. An aptamer cross-linked hydrogel as a colorimetric platform for visual detection. Angew Chem Int Ed Engl. 2010;49(6):1052–1056. doi:10.1002/anie.200905570
149. Wang Y, Shao Y, Ma X, et al. Constructing tissuelike complex structures using cell-laden DNA hydrogel bricks. ACS Appl Mater Interfaces. 2017;9(14):12311–12315. doi:10.1021/acsami.7b01604
150. Finke A, Bußkamp H, Manea M, Marx A. Designer extracellular matrix based on DNA–peptide networks generated by polymerase chain reaction. Angew Chem Int Ed Engl. 2016;55(34):10136–10140. doi:10.1002/anie.201604687
151. Zinchenko A, Che Y, Taniguchi S, Lopatina L, Sergeyev V, Murata S. Metallization of DNA hydrogel: application of soft matter host for preparation and nesting of catalytic nanoparticles. J Nanopart Res. 2016;18(7):179. doi:10.1007/s11051-016-3480-4
152. Youngblood RL, Truong NF, Segura T, Shea LD. It’s all in the delivery: designing hydrogels for cell and non-viral gene therapies. Mol Ther. 2018;26(9):2087–2106. doi:10.1016/j.ymthe.2018.07.022
153. Shahbazi M-A, Bauleth-Ramos T, Santos HA. Hydrogel Assemblies: DNA Bridging synthesis principles to biomedical applications. 2018;1(4):1800042.
154. Yang L, Yao C, Li F, Dong Y, Zhang Z, Yang D. Synthesis of branched DNA scaffolded super-nanoclusters with enhanced antibacterial performance. Small. 2018;14(16):e1800185. doi:10.1002/smll.v14.16
155. Um SH, Lee JB, Park N, Kwon SY, Umbach CC, Luo D. Enzyme-catalysed assembly of DNA hydrogel. Nat Mater. 2006;5(10):797–801. doi:10.1038/nmat1741
156. Park N, Um S H, Funabashi H, Xu J, Luo D. A Cell-free Protein-producing Gel. Vol. 82009.
157. Park N, Um SH, Funabashi H, Xu J, Luo D. A cell-free protein-producing gel. Nat Mater. 2009;8(5):432–437. doi:10.1038/nmat2419
158. Wu C, Han D, Chen T, et al. Building a multifunctional aptamer-based DNA nanoassembly for targeted cancer therapy. J Am Chem Soc. 2013;135(49):18644–18650. doi:10.1021/ja4094617
159. Yuan Q, Zhang Y, Chen T, et al. Photon-manipulated drug release from a mesoporous nanocontainer controlled by azobenzene-modified nucleic acid. ACS Nano. 2012;6(7):6337–6344. doi:10.1021/nn3018365
160. Pan YJ, Chen YY, Wang DR, et al. Redox/pH dual stimuli-responsive biodegradable nanohydrogels with varying responses to dithiothreitol and glutathione for controlled drug release. Biomaterials. 2012;33(27):6570–6579. doi:10.1016/j.biomaterials.2012.05.062
161. Kang H, Trondoli AC, Zhu G, et al. Near-infrared light-responsive core–shell nanogels for targeted drug delivery. ACS Nano. 2011;5(6):5094–5099. doi:10.1021/nn201171r
162. El-Hamed F, Dave N, Liu J. Stimuli-responsive releasing of gold nanoparticles and liposomes from aptamer-functionalized hydrogels. Nanotechnology. 2011;22(49):494011. doi:10.1088/0957-4484/22/49/494011
163. Mo R, Jiang T, Sun W, Gu Z. ATP-responsive DNA-graphene hybrid nanoaggregates for anticancer drug delivery. Biomaterials. 2015;50:67–74. doi:10.1016/j.biomaterials.2015.01.053
164. Ju C, Mo R, Xue J, et al. Sequential intra-intercellular nanoparticle delivery system for deep tumor penetration. Angew Chem Int Ed Engl. 2014;53(24):6253–6258. doi:10.1002/anie.201311227
165. Manuguerra I, Grossi G, Thomsen RP, et al. Construction of a polyhedral DNA 12-arm junction for self-assembly of wireframe DNA lattices. ACS Nano. 2017;11(9):9041–9047. doi:10.1021/acsnano.7b03538
166. Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297. doi:10.1038/nature04586
167. Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332(6027):342–346. doi:10.1126/science.1202998
168. Zhang F, Jiang S, Wu S, et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat Nanotechnol. 2015;10:779. doi:10.1038/nnano.2015.162
169. Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325(5941):725–730. doi:10.1126/science.1174251
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
