Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
STAT6 rs324015 Gene Polymorphism Increases Ulcerative Colitis Risk: A Case–Control Study
Authors Dai L, Zhang D, Qian Y, Wan Y, Chang S, Qian H
Received 9 July 2020
Accepted for publication 1 December 2020
Published 19 January 2021 Volume 2021:14 Pages 101—107
DOI https://doi.org/10.2147/PGPM.S271327
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Lingying Dai,1,* Dan Zhang,2,* Yunzhi Qian,2 Yemin Wan,2 Shuchen Chang,2 Haihua Qian2
1No. 1 Clinical Medical College, Nanjing University of Chinese Medicine, Nanjing, Jiangsu 210023, People’s Republic of China; 2Department of Colorectal Surgery, The Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, Jiangsu 210029, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Haihua Qian Tel +86-025-86617141
Fax +86-025-86617141
Email [email protected]
Introduction: Phosphorylation of signal transducer and activator of transcription 6 (STAT6) in the colonic epithelium is elevated in ulcerative colitis (UC) patients, and its inhibition prevents IL-13-associated apoptosis and barrier disruption. Recently, the STAT6 rs324015 polymorphism was reported to be related to genetic susceptibility to UC.
Methods: We examined STAT6 rs324015 using the PCR–RFLP method in 268 UC cases and 357 controls. STAT6 expression was determined by quantitative reverse-transcription PCR. The gene–environment interactions were addressed by cross-over analysis.
Results: We found that the STAT6 rs324015 polymorphism enhanced the risk of UC under the homozygous, dominant, and allelic models. Further subgroup analyses indicated that this relationship was more evident in alcohol users, smokers, and those younger than 40 years. Cross-over analysis showed strong interactions of STAT6 rs324015 with smoking/alcohol use. In addition, this polymorphism was associated with the severity, and location of UC. The GG genotype was significantly associated with increased STAT6 gene levels.
Conclusion: In summary, the STAT6 rs324015 polymorphism is related with predisposition to UC in a Chinese Han population.
Keywords: STAT6, ulcerative colitis, case–control study, rs324015 polymorphism
Introduction
Inflammatory bowel diseases (IBDs) are heterogeneous intestinal disorders, which affects multiple gastrointestinal tract organs.1 IBD reflects an imbalance between an uncontrolled inflammatory response and the intestinal microbiota.2,3 An estimated 1.5 million Americans and 2.2 million Europeans have IBD.4 There are two main types of IBD: Crohn’s disease (CD) and ulcerative colitis (UC). UC is a chronic IBD; it is characterized by chronic, relapsing inflammation process in the colorectal mucosa from the rectum to cecum. The etiology of UC is complex, and interactions among immune responses, genetic predisposition, and environmental factors contribute to its occurrence.5,6 UC causes a loss of intestinal immune tolerance due to genetic and environmental factors.7 Novel UC susceptibility loci were reported before.8,9
The signal transducer and activator of transcription 6 (STAT6) regulates Th1- and Th2-mediated immune responses.10 The intestinal mucosa showed imbalanced activation of Th2 and Th1 lymphocytes in IBD.11 STAT6 participated in IL-4- and IL-13-regulated Th2 responses.12 Li et al suggested that downregulating miR-214-3p contributed to the development of UC via targeting STAT6.13 Moreover, Rosen et al found that STAT6 deficiency ameliorated the severity of oxazolone-induced colitis by decreasing Th2-inducing cytokines.14 They also suggested that UC was associated with elevated phosphorylation of STAT6 in the colonic epithelium, and that inhibition of STAT6 prevented IL-13-induced apoptosis and barrier disruption.15 Based on the above evidence, STAT6 may play an indispensable role in the development and pathogenesis of UC. Therefore, we postulated that STAT6 may be a candidate gene for determining UC predisposition because of immunoregulatory function. The chromosomal location of STAT6 is at 12q13.3-14.1. Three studies have explored the connection between the STAT6 rs324015 polymorphism and UC predisposition, with conflicting findings.16–18 New case–control studies are required to verify previous findings; in that case, this study was designed to interpret the potential connection between STAT6 rs324015 and UC susceptibility in this Chinese Han population.
Methods
Subjects
We recruited 268 UC patients from the Affiliated Hospital of the Nanjing University of Chinese Medicine. The UC cases were diagnosed according to clinical, endoscopic, radiographic, and histological criteria.19 The severity of UC was evaluated using Truelove and Witts’ severity index, and the disease location was classified using the Montreal classification.20 The exclusion criteria were the presence of an (1) autoimmune disease, (2) malignant tumor, or (3) other known chronic inflammatory disease. The following clinical information was extracted from the medical records of the UC patients: family history, cigarette and alcohol use, and body mass index (BMI). During the same period, 357 age- and sex-matched healthy controls were recruited from healthy individuals undergoing physical examinations. Informed consents were got from all subjects.
The hospital ethics committee approved the conduction of this study; this study followed the guidelines of the Declaration of Helsinki.
Blood Sampling and Genotyping
First, blood samples (2 mL) were collected from all subjects in EDTA tubes and stored at −80°C. DNA was isolated by use of the DNA Purification Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s instructions. Genotyping was conducted by PCR–RFLP technique. The primers used were 5ʹ-GAAGTTCAGGCTCTGAGAGAC-3ʹ (forward) and 5ʹ-CCATCACCCTCAGAGAGC-3ʹ (reverse). To ensure the accuracy of genotyping, we directly sequenced 10% of the samples. The results were 100% concordant.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from PBMCs by use of the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). By use of SYBR Green I Real-Time PCR kit (GenePharma, Shanghai, China), qRT-PCR was conducted on the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). GAPDH was regarded as the internal control to normalize the expression of STAT6. The respective forward and reverse primers for PCR were 5ʹ-ATGGACAATGCCTTCTCTGA-3ʹ and 5ʹ-AACCACTGCCAAAATGTGAAC-3ʹ for STAT6 and 5ʹ-AGGTCGGTGTGAACGGATTTG-3ʹ and 5ʹ-TGTAGACCATGTAGTTGAGGTCA-3ʹ for GAPDH. The experiment was repeated three times, and the relative expression levels were determined using the 2-ΔΔCT method.
Statistical Analysis
Continuous variables were determined by utilization of the One-Way ANOVA or unpaired Student’s t-test, and the χ2 test was utilized for analyzing categorical variables. If variables did not conform to normality, Nonparametric test was employed. Hardy–Weinberg equilibrium was assessed for this polymorphism in controls using a goodness-of-fit χ2 test. Using logistic regression analyses, ORs and 95% CIs were analyzed to evaluate the connection between genetic variants and UC risk. Gene × environment interactions (including gene × smoking and gene × alcohol use) were addressed by the Cross-over analysis. Regression models were adjusted for sex and age. The power of the study was calculated at a significance level of 0.05.21 All relevant statistical analyses were conducted by use of SPSS 22.0 (SPSS, Chicago, IL, USA). P < 0.05 was considered significant.
Results
Participant Characteristics
Table 1 summarizes the demographic and risk factors of all subjects. No significant differences were shown between two groups regarding age, sex, BMI, or alcohol use. There were more smokers among the UC patients than the controls (P < 0.001). Of the UC cases, 168 had distal colitis and 100 extensive colitis. There were 111 (41.4%) mild, 125 (46.6%) intermediate, and 32 (11.9%) severe UC cases, respectively.
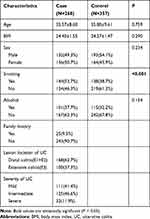 |
Table 1 Demographics of Study Participants |
Relationship Between the STAT6 rs324015 Polymorphism and UC Risk
The genotype distribution of the rs324015 polymorphism in the controls was in line with Hardy–Weinberg equilibrium (Table 2). The GG genotype of this polymorphism was correlated with UC susceptibility (GG vs AA: OR, 1.88; 95% CI, 1.02–3.45; P = 0.043). The GG and AG genotypes or G allele significantly increased the risk of UC. These findings remained significant even after adjusting for age and sex. The power analysis revealed that this study had a power of 69.6% to detect the effect of the rs324015 polymorphism on UC susceptibility, assuming an OR of 1.37.
 |
Table 2 The Association of Genotype and Allele of the STAT6 rs324015 Polymorphism with Ulcerative Colitis Risk |
We conducted stratified analyses according to sex, age, and alcohol and smoking statuses to evaluate the effect of STAT6 rs324015 on the risk of UC (Table 3). A significant association between STAT6 rs324015 and UC risk was seen in smokers, alcohol users, and subjects <40 years. However, there was no positive finding according to sex. Due to the potential interactions of smoking/alcohol use with the STAT6 rs324015 polymorphism, we used cross-over analysis to validate the results (Table 4). The GG or AG genotype was not related to the risk of UC compared with the AA genotype. Furthermore, no positive association was yielded between alcohol use and UC risk. However, we found that smokers with the GG or AG genotype presented a higher risk of UC than non-smokers with the AA genotype (GG + smoking vs AA + non-smoking: OR, 3.11, 95% CI, 1.35–7.16; P = 0.006; AG + smoking vs AA+ non-smoking: OR, 2.40, 95% CI, 1.52–3.80; P < 0.001). Alcohol drinkers with the AG genotype also showed a higher risk of UC than non-drinkers with the AA genotype. In total, this study observed interactions between environmental (alcohol and smoking) and genetic (GG or AG genotype of STAT6 rs324015) factors.
 |
Table 3 Stratified Analyses Between the STAT6 rs324015 Polymorphism and the Risk of Ulcerative Colitis |
 |
Table 4 Genetic (G) and Environmental (E) Factors 2×4 Fork Analysis |
Subsequently, we evaluated the relationships between this polymorphism and the clinical manifestations of the UC cases (Table 5). We found some genotypes of rs324015 polymorphism were related with the severity, and lesion location of UC.
 |
Table 5 The Association Between the STAT6 rs324015 Polymorphism and Clinical Characteristics of Ulcerative Colitis |
Last, we evaluated the impact of the STAT6 rs324015 polymorphism on STAT6 gene expression. Compared with the AA genotype, the GG genotype was significantly associated with increased STAT6 expression (P = 0.034; Figure 1).
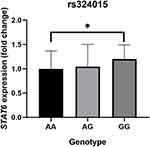 |
Figure 1 The relative STAT6 mRNA expression among three genotypes of rs324015 polymorphism (*P < 0.05, GG genotype vs AA genotype). |
Discussion
In this present case-control study, we observed that the STAT6 rs324015 polymorphism enhanced the risk of UC in Chinese individuals. Subgroup analyses indicated that the STAT6 rs324015 was related to an increased risk of UC in smokers, alcohol users, and those individuals under 40 years. STAT6 rs324015 was linked with the location, and severity of UC, and the GG genotype was significantly associated with increased STAT6 expression.
Xia et al first explored the relationship between the rs324015 polymorphism of STAT6 gene and IBD risk in a Dutch population.16 They showed that UC did not differ according to the genetic distribution when it was classified by disease extent, age of onset, and colectomy, indicating that STAT6 rs324015 polymorphism did not confer susceptibility to UC.16 A German study by Klein et al revealed that the G allele and GG genotype were significantly related with an elevated risk of CD only, but not of UC.17 A subsequent Chinese study suggested that STAT6 rs324015 was not associated with genetic susceptibility to UC in Chinese patients.18 However, unlike the abovementioned studies,16–18 we uncovered that this polymorphism elevated the risk of UC. We postulate the following reasons for these inconsistent findings. First, the allele and genotype distributions of STAT6 rs324015 differed significantly between our cohort and the other study populations. Second, the exposure factors for UC were diverse among the studies. Third, UC is clinically heterogeneous; for example, the severities and lesion locations of UC are distinct. Fourth, the sample sizes of these studies varied.
In further analyses, we evaluated the relationships between STAT6 rs324015 and environmental factors. This polymorphism was linked with an enhanced risk of UC in smokers, alcohol users, and those aged lower than 40 years, suggesting that interactions of smoking, alcohol use, and STAT6 rs324015 may account for an increased risk of UC in this population. However, no significant association between the rs324015 polymorphism and sex was seen. Due to the potential interactions of smoking/alcohol use with the STAT6 rs324015 polymorphism, cross-over analysis was conducted. Significant interactions between genetic factors (GG or AG genotype of STAT6 rs324015) and alcohol use/smoking were detected. Next, we investigated the connection between the STAT6 rs324015 polymorphism and the clinical manifestations of the UC patients. This polymorphism was related to the severity, and location of UC. The data suggest that the rs324015 polymorphism confers increased susceptibility to UC in those with extensive colitis or severe UC.
The following study limitations should be noted. First, the sample size was small, which may reduce the reliability of the results. Second, selection bias existed in this hospital-based case-control study; however, we could not predict the impact of selection bias on the results. Third, the limited data restricted further analyses. Fourth, evaluation of only one polymorphism is insufficient because rs324015 may be in linkage disequilibrium with other polymorphisms. Fifth, immunological indicators such as IL-4, IL-13, Th1 and Th2 lymphocytes should be tested. Finally, other genetic–environmental interactions that were not assessed may exist.
Conclusion
The STAT6 rs324015 polymorphism is related with an enhanced risk of UC in Chinese Han individuals and is also associated with STAT6 expression. Further studies of this polymorphism in larger sample sizes are needed.
Author Contributions
All authors contributed to conception and design, acquisition of data, or analysis and interpretation of data substantially; were involved in drafting this paper or revising it critically; were in agreement with submitting to the current journal; approved the version to be published; and agree to be responsible for all aspects of this study.
Funding
There is no funding to report.
Disclosure
The authors report no conflict of interest.
References
1. Ganji-Arjenaki M, Rafieian-Kopaei M. Phytotherapies in inflammatory bowel disease. J Res Med Sci. 2019;24:42. doi:10.4103/jrms.JRMS_590_17
2. de Mattos BR, Garcia MP, Nogueira JB, et al. Inflammatory bowel disease: an overview of immune mechanisms and biological treatments. Mediators Inflamm. 2015;2015:493012. doi:10.1155/2015/493012
3. Mizoguchi A, Takeuchi T, Himuro H, Okada T, Mizoguchi E. Genetically engineered mouse models for studying inflammatory bowel disease. J Pathol. 2016;238(2):205–219. doi:10.1002/path.4640
4. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12(4):205–217. doi:10.1038/nrgastro.2015.34
5. Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi:10.1038/nature11582
6. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389(10080):1756–1770. doi:10.1016/S0140-6736(16)32126-2
7. Uhlig HH, Muise am. clinical genomics in inflammatory bowel disease. Trends Genet. 2017;33(9):629–641. doi:10.1016/j.tig.2017.06.008
8. McGovern DP, Kugathasan S, Cho JH. Genetics of inflammatory bowel diseases. Gastroenterology. 2015;149(5):1163–1176 e1162. doi:10.1053/j.gastro.2015.08.001
9. Upadhyay R, Dua B, Sharma B, et al. Transcription factors STAT-4, STAT-6 and CREB regulate Th1/Th2 response in leprosy patients: effect of M. leprae antigens. BMC Infect Dis. 2019;19(1):52. doi:10.1186/s12879-018-3601-z
10. Van Damme N, De Keyser F, Demetter P, et al. The proportion of Th1 cells, which prevail in gut mucosa, is decreased in inflammatory bowel syndrome. Clin Exp Immunol. 2001;125(3):383–390. doi:10.1046/j.1365-2249.2001.01638.x
11. Shimoda K, van Deursen J, Sangster MY, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380(6575):630–633. doi:10.1038/380630a0
12. Li JA, Wang YD, Wang K, et al. Downregulation of miR-214-3p may contribute to pathogenesis of ulcerative colitis via targeting STAT6. Biomed Res Int. 2017;2017:8524972. doi:10.1155/2017/8524972
13. Rosen MJ, Chaturvedi R, Washington MK, et al. STAT6 deficiency ameliorates severity of oxazolone colitis by decreasing expression of claudin-2 and Th2-inducing cytokines. J Immunol. 2013;190(4):1849–1858. doi:10.4049/jimmunol.1201373
14. Rosen MJ, Frey MR, Washington KM, et al. STAT6 activation in ulcerative colitis: a new target for prevention of IL-13-induced colon epithelial cell dysfunction. Inflamm Bowel Dis. 2011;17(11):2224–2234. doi:10.1002/ibd.21628
15. Xia B, Crusius JB, Wu J, Zwiers A, van Bodegraven AA, Pena AS. Signal transducer and activator of transcription 6 gene G2964A polymorphism and inflammatory bowel disease. Clin Exp Immunol. 2003;131(3):446–450. doi:10.1046/j.1365-2249.2003.02079.x
16. Klein W, Tromm A, Folwaczny C, et al. The G2964A polymorphism of the STAT6 gene in inflammatory bowel disease. Dig Liver Dis. 2005;37(3):159–161. doi:10.1016/j.dld.2004.10.011
17. Zhu J, Xia B, Guo Q, et al. Distribution of signal transducer and activator of transcription 6 gene G2964A polymorphism in Chinese patients with ulcerative colitis. J Gastroenterol Hepatol. 2006;21(12):1854–1857. doi:10.1111/j.1440-1746.2006.04427.x
18. Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170(2–6):discussion 16–19.
19. Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55(6):749–753. doi:10.1136/gut.2005.082909
20. Hedges LV, Pigott TD, The power of statistical tests in meta-analysis. Psychol Methods. 2001;6(3):203–217. doi:10.1037/1082-989X.6.3.203
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
