Back to Journals » OncoTargets and Therapy » Volume 8
SOX2 expression is associated with FGFR fusion genes and predicts favorable outcome in lung squamous cell carcinomas
Authors Zheng S, Pan Y, Wang R , Li Y, Cheng C, Shen X, Li B, Zheng D, Sun Y, Chen H
Received 27 June 2015
Accepted for publication 10 September 2015
Published 19 October 2015 Volume 2015:8 Pages 3009—3016
DOI https://doi.org/10.2147/OTT.S91293
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Shanbo Zheng,1,2,* Yunjian Pan,1,2,* Rui Wang,1,2 Yuan Li,2,3 Chao Cheng,1,2 Xuxia Shen,2,3 Bin Li,1,2 Difan Zheng,1,2 Yihua Sun,1,2 Haiquan Chen1,2
1Department of Thoracic Surgery, Fudan University Shanghai Cancer Center, Shanghai, People’s Republic of China; 2Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, People’s Republic of China; 3Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Objectives: SOX2 is a gene that encodes for a transcription factor, which functions as an activator or suppressor of gene transcription. SOX2 amplification and overexpression have been found in various types of tumors and play important roles in cancer cells. The aim of the study was to evaluate SOX2 expression and amplification in lung squamous cell carcinomas (SCCs) and to determine the relationship with main clinicopathologic features, patient prognosis, and common driver mutations.
Materials and methods: SOX2 protein levels were measured by immunohistochemistry, while SOX2 copy numbers were measured by fluorescence in situ hybridization in resected samples from 162 Chinese lung SCC patients. All patients were also analyzed for mutations in EGFR, HER2, BRAF, PIK3CA, NFE2L2, and FGFR fusion genes. Clinical characteristics, including age, sex, smoking status, stage, relapse-free survival (RFS), and overall survival (OS), were collected.
Results: SOX2 overexpression and amplification were observed in 58.6% and 45.9% of lung SCCs. Lung SCC patients with SOX2 overexpression were significantly associated with absence of malignant tumor family history (P=0.021), FGFR fusion gene (P=0.046), longer RFS (P=0.041), and OS (P=0.025). No correlation was found between SOX2 gene amplification and main clinicopathologic features, patient prognosis, or common driver mutations.
Conclusion: SOX2 overexpression and amplification are common in lung SCCs. SOX2 overexpression was associated with FGFR fusion genes and predicted favorable outcome in lung SCCs. The underlying relationship of SOX2 and FGFR still needs further investigation.
Keywords: lung squamous cell carcinoma, SOX2 amplification, protein expression, FGFR fusion gene, prognostic marker
Introduction
Lung cancer continues to be the leading cause of cancer-related deaths worldwide.1 Despite multidisciplinary cancer therapies having taken great strides during the past decade, the overall prognosis for lung cancer patients remains poor. In lung adenocarcinoma, small molecule inhibitors targeting activated EGFR and EML4-ALK have improved the response rates and progression-free survival;2,3 however, lung squamous cell carcinomas (SCCs) lack novel treatment strategies that target molecular abnormalities. Therefore, it is urgently needed to identify reliable prognostic biomarkers and develop targeted molecular therapies for lung SCCs.
SOX2 (SRY [sex-determining region Y] box 2) is a recently identified novel lineage-survival oncogene in SCCs, which is located on chromosome 3q26.33.4 The SOX2 protein is a transcription factor of 317 amino acids containing a high mobility group domain.5 It plays a crucial role in both the pluripotency regulation in embryonic stem cells and the morphogenesis and homoeostasis of tracheobronchial epithelia.6 Moreover, SOX2 amplification and protein expression have been found in various types of tumors,7–11 and recent studies have shown that SOX2 is responsible for cellular proliferation, tumor invasion and migration, self-renewal, and maintenance in cancer stem cell populations.12 Amplified and overexpressed SOX2 is frequently associated with higher tumor-node-metastasis (TNM) stage, metastasis, and poorer prognosis in SCCs,13–15 but with opposing consequences of better outcome in lung carcinomas.16,17 The conflicting results suggest that the role of SOX2 in the clinic is still not well defined. Furthermore, the relationship between SOX2 and common molecular abnormalities in lung SCCs is still largely unexplored.
In this study, we performed fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) in surgically resected lung SCC and correlated amplification and expression of SOX2 with the main clinicopathologic features, patient prognosis, and common driver mutations.
Materials and methods
Patients and samples
Lung SCCs in the form of formalin-fixed paraffin embedded tissue from patients who underwent surgical resection with curative intent at Fudan University Shanghai Cancer Center between January 2008 and December 2011 were obtained. Eligible patients were required to have sufficient tissue for immunohistochemical staining, FISH, and comprehensive mutational analyses. Patients who received neoadjuvant chemotherapy or had a history of malignant tumor were excluded.
Clinicopathologic variables collected for analyses included sex, age at diagnosis, smoking history, family history of malignant tumor, tumor differentiation, pathologic TNM stage in line with the seventh edition of the lung cancer staging system,18 and postoperative adjuvant chemotherapy/radiotherapy. Disease recurrence and survival were observed in the follow-up clinic or obtained via telephone. The average follow-up period was 40 months (median 42 months range [2, 88]). All patients provided written informed consent at the interview. This study was approved by the Committee for Ethical Review of Research (Fudan University Shanghai Cancer Center IRB#090977-1).
Immunohistochemistry
In brief, sections were deparaffinized by serial xylene washes and rehydrated in graded alcohols and were then treated with 3% H2O2 to block endogenous peroxidase activity. Antigen retrieval was done by immersing slides in citrate buffer (pH 6) followed by microwaving. Nonspecific immunoglobulin binding was blocked using 10% goat serum in phosphate-buffered saline (PBS) (Sigma-Aldrich, St Louis, MO, USA). The slides were incubated with primary SOX2 rabbit monoclonal antibody (clone D6D9; Cell Signaling Technology, Danvers, MA, USA) diluted 1:100 in SignalStain antibody diluent (Cell Signaling Technology) overnight at 4°C. After incubation with the primary antibody, sections were washed with PBS and incubated with secondary antibodies, which was followed by incubation with 3,3′-diaminobenzidine. The slides were then counterstained with hematoxylin.
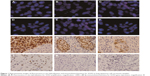
The expression level of SOX2 was measured independently by two pathologists (Yuan Li and Xuxia Shen) who were blinded to the clinical data, and discrepancies were resolved by reviewing the corresponding sections and discussion. Only nuclear SOX2 expression was evaluated. Nuclear SOX2 immunohistochemical staining was quantified using a four-value intensity score (0, 1+, 2+, and 3+) and the percentage (0%–100%) of the reactivity extension. The final score was then obtained by multiplying the intensity and reactivity extension values (range, 0–300). A score of 100 was used as the cutoff value between positive and negative protein expression. Representative images of staining intensities of SOX2 are shown in Figure 1.
Fluorescence in situ hybridization
FISH analysis of SOX2 amplification was performed using a spectrum orange-labeled probe CTD-2348H10 and SPOT-Light tissue pretreatment solution (Invitrogen, Carlsbad, CA, USA). Briefly, sections were deparaffinized by serial xylene washes and rehydrated in graded alcohols, then digested with protease K (0.5 mg/mL) at 37°C for 20 minutes. The slides were then dehydrated in ethanol. The probes were denatured for 5 minutes at 75°C before hybridization. Slides were hybridized at 37°C for 36 hours and washed in 2× SSC/0.3% NP-40 at 72°C for 2 minutes. Nuclei were counterstained with DAPI 1 counterstain (Vysis, Downers Grove, IL, USA). Analyses were performed using a fluorescence microscope (Olympus BX51TRF, Olympus Corporation, Tokyo, Japan) equipped with an Olympus BX-UCB filter set (Olympus) with single-band exciters for Texas red/rhodamine, fluorescein isothiocyanate, and DAPI (UV 360 nm). Copy number per cell for each gene was enumerated on at least 50 tumor cells for each case. In the absence of validated FISH scoring criteria for SOX2, a cutoff value of more than 4 gene copies/cell, or presence of gene clusters, was set to identify cases with increased gene copy number (FISH+). Examples of FISH patterns are shown in Figure 1.
Mutational analyses
Comprehensive mutational analyses of EGFR, HER2, BRAF, FGFR, PIK3CA, and NFE2L2 were performed in lung SCCs. In brief, frozen tissues were dissected into TRIzol (Life Technologies, Carlsbad, CA, USA) followed by total RNA extraction using standard protocol. Total RNA samples were reverse transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Fermentas, St Leon-Rot, Germany). EGFR (exons 18–22), HER2 (exons 18–21), BRAF (exons 11–15), FGFR (FGFR1, FGFR2, and FGFR3), PIK3CA (exons 9 and 20), and NFE2L2 (exon 2) were amplified by polymerase chain reaction using cDNA. Amplified products were analyzed by direct dideoxynucleotide sequencing.
Statistical analyses
Difference in proportions was analyzed by Pearson’s chi-square test or Fisher’s exact test. Relapse-free survival (RFS) and overall survival (OS) of patients with positive or negative IHC and FISH were estimated using the Kaplan–Meier method. The log-rank test was used to determine survival differences between groups. Independent prognostic factors were identified through the Cox proportional hazards regression (forward likelihood ratio model). All tests were two-tailed. Statistical significance was set as P<0.05. All data were analyzed using the SPSS Version 19.0 Software (SPSS Inc., Chicago, IL, USA).
Results
Patient characteristics
A total of 162 lung SCC samples were collected from 12 females and 150 males. The mean age of the patients was 61.6 years, ranging from 40 to 88 years. One hundred thirty-eight patients (85.2%) had a history of smoking, and 36 patients (22.2%) had a family history of malignant tumor. The mean diameter of tumor was 4.306 cm, ranging from 0.7 to 13.0 cm. About 68 (42.0%) patients presented with lymph node metastasis. The patient characteristics of this cohort are described in Table 1.
  | Table 1 Patient characteristics |
Correlation between SOX2 expression and clinicopathologic and molecular features
SOX2 expression status was successfully determined in 145 patients, and positive staining was observed in 85 tumors (Table 2). SOX2 overexpression was not associated with age, sex, smoking history, tumor differentiation, and pleural invasion. Positive SOX2 expression was significantly associated with absence of malignant tumor family history (P=0.021). Although there was a trend that SOX2-positive patients had smaller tumor diameter (4.557 cm vs 4.021 cm, P=0.115) and less advanced pathologic stage (P=0.268), the differences did not reach a statistical significance. Interestingly, among the six detected common molecular abnormalities in non-small-cell lung cancer (NSCLC), a statistically significant association was observed between SOX2 expression and FGFR fusion gene. Of 162 samples, FGFR3-TACC3 was identified in 8 patients and BAG4-FGFR1 was found in 2 patients. We found that seven patients with FGFR3-TACC3 and two with BAG4-FGFR1 were positive for SOX2 staining and only one patient with FGFR3-TACC3 was negative for SOX2 expression (P=0.046).
  | Table 2 Clinicopathologic and molecular characteristics according to SOX2 expression and amplification |
Correlation between SOX2 amplification and clinicopathologic and molecular features
We also examined SOX2 amplification using FISH in 111 cases. SOX2 amplification was detected in 50 sections, and SOX2 gene copy number ranged from 2.2 to 15.3 (mean, 4.46). No correlation was found between SOX2 gene amplification in SCC and the patient’s age, sex, smoking history, family history of malignant tumor, tumor diameter, tumor-infiltrating lymphocytes, pathologic stage, pleural invasion, and mutational status.
Our results revealed that SOX2 FISH results were correlated with SOX2 IHC results (Table 3, Spearman correlation coefficient =0.453; P<0.001). Altogether 94 samples were examined for both SOX2 expression and amplification. Of 40 SOX2 FISH positive cases, 33 (82.5%) showed positive SOX2 IHC and only 7 cases (17.5%) were SOX2 IHC negative. Of 54 SOX2 FISH negative cases, 34 cases (63.0%) were negative for SOX2 IHC, and 20 cases (37.0%) were SOX2 IHC positive.
  | Table 3 Comparison of SOX2 amplification and expression |
Survival analysis
The log-rank test on the Kaplan–Meier survival analysis demonstrated that lung SCC patients with positive SOX2 expression had longer RFS (Figure 2, median survival 55 vs 36 months; log rank P=0.041) and OS (Figure 2, median survival 65 vs 44 months; log rank P=0.025) than subjects with negative SOX2 expression. In contrast, although the median RFS and OS time for patients with SOX2 FISH positive lung SCC were better than SOX2 FISH negative patients, the differences were not statistically significant (Figure 2, for RFS, median survival 43 vs 38 months, log rank P=0.630; for OS, median survival 55 vs 43 months, log rank P=0.141).
To determine the prognostic accuracy of SOX2 expression, we used multivariate Cox regression model adjusted for sex, age at diagnosis, smoking history, family history of malignant tumor, tumor differentiation, TNM stage, mutational status, and postoperative chemotherapy/radiotherapy. However, Cox proportional hazards regression analysis revealed that SOX2 protein expression was associated with age and TNM stage, and failed to demonstrate SOX2 protein expression to be an independent prognostic factor in lung SCC.
Discussion
This study evaluated the relationship of clinical pathologic features, patient prognosis, and common driver mutations with protein expression and copy number alterations of SOX2 in a cohort of patients with surgically resected lung SCC. For the first time, we report that high level SOX2 protein expression assessed by IHC is associated with FGFR fusion gene in lung SCC.
The correlation of SOX2 expression with clinicopathological characteristics has been investigated in several retrospective NSCLC series. High-level expression of SOX2 was reported to be associated with lower TNM grade, smaller tumor size, lower probability of invasion and metastasis, and former or current smoking history,19,20 but recently a meta-analysis showed no correlation between SOX2 expression and clinicopathological parameters such as age, sex, smoking, lymph node metastasis, and tumor stage.17 Here, our results showed that SOX2 positive expression was associated with absence of malignant tumor family history. Our new results showed the probability that SOX2 overexpression has less relationship with genetic factors, but more association with environmental impacts, and the regulation of SOX2 protein remains to be further explored.
Recently, the prognostic role of SOX2 expression and amplification in NSCLC has been investigated in different studies,16,19–25 but the results were contradictory. Several studies associated SOX2 protein overexpression with prolonged survival in surgically resected lung SCC patients,19,21,25 but a poor outcome was shown in early stage lung adenocarcinomas in the study of Sholl et al.24 Meanwhile, Luca et al23 reported increased SOX2 gene copy number as an independent favorable prognostic factor in patients with stage I and II NSCLC, but Brcic et al25 showed a nonsignificant result. Here, we carried out a study in a Chinese cohort and proved that SOX2 protein overexpression predicts better RFS and OS in lung SCC, but is not an independent prognostic factor.
SOX2 has shown its potential not only to become a useful biomarker for prognosis in the clinic, but also to be a novel therapy option. Chen et al26 demonstrated that silencing of the SOX2 gene effectively induced apoptosis via the activation of death receptor and mitochondrial signaling pathways in human NSCLC cells. Dogan et al27 investigated that SOX2 knockdown using shRNA in lung adenocarcinoma cell lines decreased cell proliferation and increased cell sensitivity to erlotinib. Moreover, SOX2 expression decreased when treated with PI3K/AKT inhibitors. Therefore, utilizing SOX2 and its upstream or downstream proteins for cancer therapy could open a window to new therapeutic opportunities, and identification of the relationship between SOX2 protein expression or gene amplification and common driver genes may help to identify patients who might benefit from particular targeted therapies.
FGFR fusion gene was illustrated to be a new driver for a range of cancers.28 A previous study has reported that FGFR1/3 fusions occurred in 1.3% of patients with NSCLCs and in 3.5% of patients with lung SCC.29 FGFR fusions have been shown to sensitize cancer cells to FGFR kinase inhibitors PD173074 and pazopanib,30 suggesting that a new subset of cancers may be treatable with FGFR-targeted therapy. To our knowledge, this study was the first to represent the association of SOX2 gene expression with FGFR fusion in a clinical cohort. Although the underlying mechanism between SOX2 and FGFR still needs to be further developed, novel therapies targeting FGFR or even SOX2 may be promising.
Conclusion
In conclusion, our data demonstrated that positive SOX2 expression was significantly associated with absence of malignant tumor family history, FGFR fusion genes and had a favorable clinical outcome in lung SCC patients. Our finding emphasized the importance of SOX2 in lung SCC biology and encourages further exploration of novel therapeutic combinations.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. | ||
Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. | ||
Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. | ||
Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. | ||
Adameyko I, Lallemend F, Furlan A, et al. Sox2 and Mitf cross-regulatory interactions consolidate progenitor and melanocyte lineages in the cranial neural crest. Development. 2012;139:397–410. | ||
Hussenet T, Dali S, Exinger J, et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One. 2010;5:e8960. | ||
Hussenet T, du Manoir S. SOX2 in squamous cell carcinoma: amplifying a pleiotropic oncogene along carcinogenesis. Cell Cycle. 2010;9:1480–1486. | ||
Maier S, Wilbertz T, Braun M, et al. SOX2 amplification is a common event in squamous cell carcinomas of different organ sites. Hum Pathol. 2011;42:1078–1088. | ||
Freier K, Knoepfle K, Flechtenmacher C, et al. Recurrent copy number gain of transcription factor SOX2 and corresponding high protein expression in oral squamous cell carcinoma. Genes Chromosomes Cancer. 2010;49:9–16. | ||
Andreas S, Maike B, Friederike J, et al. Expression and role of the embryonic protein SOX2 in head and neck squamous cell Carcinoma. Carcinogenesis. 2014;35(7):1636–1642. | ||
Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med. 2014;3:19. | ||
Honing J, Pavlov KV, Meijer C, et al. Loss of CD44 and SOX2 expression is correlated with a poor prognosis in esophageal adenocarcinoma patients. Ann Surg Oncol. 2014;21:657–664. | ||
Han X, Fang X, Lou X, et al. Silencing SOX2 induced mesenchymal-epithelial transition and its expression predicts liver and lymph node metastasis of CRC patients. PLoS One. 2012;7:e41335. | ||
Tian Y, Jia X, Wang S, et al. SOX2 oncogenes amplified and operate to activate AKT signaling in gastric cancer and predict immunotherapy responsiveness. J Cancer Res Clin Oncol. 2014;140:1117–1124. | ||
Velcheti V, Schalper K, Yao X, et al. High SOX2 levels predict better outcome in non-small cell lung carcinomas. PLoS One. 2013;8:e61427. | ||
Chen Y, Huang Y, Huang Y, et al. The prognostic value of SOX2 expression in non-small cell lung cancer: a meta-analysis. PLoS One. 2013;8:e71140. | ||
Detterbeck FC, Boffa DJ, Tanoue LT. The new lung cancer staging system. Chest. 2009;136(1):260–271. | ||
Wilbertz T, Wagner P, Petersen K, et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol. 2011;24:944–953. | ||
Yuan P, Kadara H, Behrens C, et al. Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PLoS One. 2010;5:e9112. | ||
Sasaki H, Yokota K, Hikosaka Y, et al. Increased Sox2 copy number in lung squamous cell carcinomas. Exp Ther Med. 2012;3:44–48. | ||
Li X, Wang J, Xu Z, et al. Expression of Sox2 and Oct4 and their clinical significance in human non-small-cell lung cancer. Int J Mol Sci. 2012;13:7663–7675. | ||
Luca T, Giovanna F, Teresa TN, et al. Increased SOX2 gene copy number is associated with FGFR1 and PIK3CA gene gain in non-small cell lung cancer and predicts improved survival in early stage disease. PLoS One. 2014;9(4):e95303. | ||
Sholl LM, Barletta JA, Yeap BY, et al. Sox2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. Am J Surg Pathol. 2010;34:1193–1198. | ||
Brcic L, Sherer CK, Shuai Y, et al. Morphologic and clinicopathologic features of lung squamous cell carcinomas expressing Sox2. Am J Clin Pathol. 2012;138:712–718. | ||
Chen S, Li X, Lu D, et al. SOX2 regulates apoptosis through MAP4K4-survivin signaling pathway in human lung cancer cells. Carcinogenesis. 2014;35(3):613–623. | ||
Dogan I, Kawabata S, Bergbower E, et al. SOX2 expression is an early event in a murine model of EGFR mutant lung cancer and promotes proliferation of a subset of EGFR mutant lung adenocarcinoma cell lines. Lung Cancer. 2014;85(1):1–6. | ||
Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling – a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–1862. | ||
Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res. 2014;20(15):4107–4114. | ||
Sabnis AJ, Bivona TG. FGFR fusions in the driver’s seat. Cancer Discov. 2013;3:607–609. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.