Back to Journals » OncoTargets and Therapy » Volume 11
Sirt3 enhances glioma cell viability by stabilizing Ku70–BAX interaction
Authors Luo K, Huang W, Tang S
Received 30 April 2018
Accepted for publication 12 July 2018
Published 29 October 2018 Volume 2018:11 Pages 7559—7567
DOI https://doi.org/10.2147/OTT.S172672
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Ke Luo,* Wei Huang,* Shuang Tang
Department of Neurosurgery, Suining Central Hospital, Suining, Sichuan, China
*These authors contributed equally to this work
Background: As one of the most prevalent malignancies, glioma is characterized by poor prognosis and high mortality rate. Glioma patients may show completely distinct clinical outcomes due to their different molecular patterns. Sirtuin 3 (Sirt3) participates in aging, stress resistance, and metabolic regulation. Here we aimed to test the expression and function of Sirt3 in glioma.
Methods: We enrolled 114 patients and tested the protein level of Sirt3 in their glioma tissues. The correlation between prognosis and Sirt3 was evaluated by univariate and multivariate analyses. We also conducted cellular experiments in U87 and U251 glioma cells, including overexpression and knockdown assays.
Results: Sirt3 expression was lower in glioma tissues than normal brain tissues. Higher Sirt3 is significantly correlated to advanced tumor grade (P=0.004), showing its potential role in cancer progression. Consistently, univariate and multivariate analyses identified Sirt3 as an independent prognostic factor (P=0.017). Patients with higher Sirt3 expression showed significantly shorter overall survival time. Moreover, overexpression of Sirt3 in either cell line enhanced cell viability, while silencing Sirt3 attenuated cell growth. Molecular assays showed Sirt3 can deacetylate Ku70 protein, therefore stabilizing the Ku70-BAX interaction. Since Ku70 can help prevent BAX transporting into mitochondria and decrease cell apoptosis, Sirt3 protein may play roles in maintaining cell viability. In addition, silencing Ku70 inhibited the pro-proliferative effect by Sirt3.
Conclusion: Taken together, our results not only identified the prognostic role of Sirt3 in glioma patients but also provided signaling pathway evidence for its functioning mechanisms.
Keywords: glioma, prognosis, Sirt3, viability
Introduction
Primary glioma is the most common type of neoplasm in the central nervous system.1 Gliomas are divided into four grades (I–IV) according to the WHO classification.2 The treatment methods toward glioma include chemotherapy, radiotherapy, and surgical intervention.3 However, the curative effect varies among individuals. Although the patients with WHO grade I–II have a better overall survival, the 5-year survival rate for grade IV patients is less than 5%.4 The pathogenesis and progression mechanisms of glioma have drawn great attention in the past decades; however, the overall improvement in patients’ outcomes is far from satisfying.5 In addition, the current prediction models on patients’ prognosis are largely based on the clinicopathological characteristics, while the efficiency varies in different studies. Therefore, a more detailed stratified approach based on molecular expression patterns is still one of the major challenges.
Sirtuins are a class of protein deacetylases and are highly conserved in many species.6 Sirtuins consist of seven protein members from sirtuin 1 to sirtuin 7.7 Different sirtuin subtypes show completely distinct subcellular localizations and functions. As for sirtuin 3 (Sirt3), it is initially characterized as a mitochondrial deacetylase involved in respiratory chain, tricarboxylic acid cycle, and fatty acid oxidation.8 Subsequently, the expression of Sirt3 was also detected in cytoplasm and nucleus with novel substrates.9 From the physiological aspect, Sirt3 participates in regulating cardiovascular functions, aging, and metabolic process.10 Most recently, Sirt3 was reported to function in tumor progression.11 For example, it inhibits tumor progression of breast cancer, hepatocellular carcinoma, and leukemia.12–14 However, the role of Sirt3 seems more like tumor promoter in colon cancer, gastric cancer, melanoma, and renal cancer.15–18 Therefore, Sirt3 can play both tumor-suppressing and tumor-promoting roles in different tumor types.
Here we initially investigated the expression and function of Sirt3 in glioma and described the mechanisms by which it modulates glioma progression. Briefly, Sirt3 was identified to be expressed higher in glioma tissues than normal brain tissues, which was significantly correlated to advanced tumor stages. Besides, the higher Sirt3 protein levels indicated poorer overall survival of glioma patients. Cellular experiments were performed in two glioma cell lines; both demonstrated the possible signaling axis of Sirt3–Ku70–BAX (Bcl-2-associated X protein) in modulating glioma progression.
Methods
Patients and samples
This study was approved by the Ethics Committee of Suining Central Hospital. Written informed consents were obtained from all patients. Formalin-fixed paraffin-embedded glioma tissues were obtained from surgery in 114 randomly selected patients during 2002–2012 in Suining Central Hospital. Another 22 glioma samples, which were fresh frozen in liquid nitrogen, were also collected in our hospital. We also purchased 17 normal brain tissue samples from Cureline (South San Francisco, CA, USA). All patients were followed-up ranging from 8 to 71 months.
Immunohistochemistry (IHC) and IHC evaluation
As described by others,19 IHC was used to evaluate the Sirt3 protein expression in glioma tissues. Briefly, deparaffinized and rehydrated slides were treated in 10 mM citrate buffer (pH 6.0) at 95°C for antigen recovery, followed by quenching the endogenous peroxidase activity with 3% H2O2 incubation. Five percent FBS was used for blocking the nonspecific binding sites. Slides were then incubated with primary antibody (anti-Sirt3; Abcam, Cambridge, MA, USA; #ab86671; 1:200) at 4°C overnight. DAB staining kit (Tiangen, China) was used to detect the immunoreactivity according to the manufacturer’s instructions.
The level of Sirt3 was determined by the degree of staining intensity and the percentage of positively stained cells. Examination and scoring were performed by two pathologists independently. In brief, weak staining, moderate staining, and strong staining were scored as 1, 2, and 3, respectively. The percentages of positively stained cells were scored as follows: 1 for less than 20%, 2 for 20%–50%, and 3 for 50%–100%. The final IHC score was defined by multiplying the the two scores above, ranging from 1 to 9. Tissues with a final score no more than 4 were regarded as low-expression cases; otherwise, it will be grouped into the Sirt3 high-expression group.
Quantitative real-time PCR (RT-qPCR)
Total RNA was extracted from fresh-frozen specimens by using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) following a standard protocol.20 Then the RNA was reversely transcribed by using the Primer-Script RT Enzyme Mix. RT-qPCR was carried out using the SYBR Premix Ex Tag (Takara, Japan) according to the instructions. GAPDH was used as normalization control, and the primers were presented as following:
Sirt3-F: 5′-ACCCAGTGGCATTCCAGAC-3′; |
Sirt3-R: 5′-GGCTTGGGGTTGTGAAAGAAG-3′; |
GAPDH-F: 5′-AGGGCTGCTTTTAACTCTGGT-3′; |
GAPDH-R: 5′-CCCCACTTGATTTTGGAGGGA-3′. |
Cell culture and transfection
Normal human astrocytes (NHA cells) were purchased from Lonza (Basel, Switzerland). The human glioma cells (U87 and U251) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM medium supplemented with 10% FBS (Hyclone, South Logan, UT, USA) and 1% streptomycin and penicillin in a humidified atmosphere of 5% CO2 at 37°C. The Sirt3 siRNA (sequence: CATCCCTACATGCAGATGAAA), Ku70 siRNA (sequence: GAGUGAAGAUGAGUUGACA), Sirt3 overexpressing plasmid construct, and Ku70 overexpressing plasmid construct used in this study were synthesized by Genechem (Shanghai, China). The transfections were carried out using Lipofectamine 3000 according to the manufacturer’s procedure (Invitrogen).21
Western blotting
Cells were scraped with PBS and lysed with ice-cold RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) supplemented with protease inhibitor cocktail. The lysates were centrifuged at 12,000× g for 15 minutes at 4°C, and the precipitate was discarded. Equal amount protein (10 μg) was electrophoresed with sodium dodecyl sulfate-polyacrylamide gel and then the proteins were transferred to a polyvinylidene difluoride membrane (Merck Millipore, Kenilworth, NJ, USA). The primary antibodies used in this study included anti-Sirt3 (Abcam, #ab86671), anti-Ku70 (Invitrogen, #MA5-13110), anti-acetyl (Abcam, #ab80178), anti-BAX (Cell Signaling Technology, Danvers, MA, USA; #2774), anticytochrome c (Cell Signaling Technology, #4280), anticaspase 3 (Cell Signaling Technology, #9662), and anti-β-actin (Cell Signaling Technology, #3700). After incubation with primary and secondary antibodies, the immunoreactivity bands were visualized by using an enhanced chemiluminescence substrate kit (Thermo Scientific, Pittsburgh, PA, USA).22
Immunoprecipitation
To identify the interaction between Sirt3 and Ku70, we performed antibody-affinity immunoprecipitation in U251 cells transfected with Ku70 plasmid. Briefly, transfected cells were harvested and lysed in lysis buffer. After centrifuged at 12,000× g at 4°C for 15 minutes, supernatants were added with Ku70–Protein G mixture (Santa Cruz Biotechnology) and incubated overnight at 4°C. Protein G was then spinned down and washed with lysis buffer for three times.23 Finally, the immunoprecipitated proteins were subjected to Western blot analyses.
Mitochondrial fractionation
To determine the protein distribution in mitochondria, cultured cells were harvested for mitochondrial fractionation by using a mitochondrial isolation kit for mammalian cells (Thermo Scientific) according to the manufacturer’s protocol.24
Cell viability assay
To evaluate the effect of Sirt3 on tumor cell viability, plasmid or siRNA transfected cells were seeded at 2×104 cells per well in a 96-well plate and cultured in DMEM. At designated time points, cell viability was assessed by a Cell Counting Kit-8 (Dojindo, Japan) according to the manufacturer’s instructions.25 Briefly, 10 μL of CCK-8 reagent was added into each well and incubated for 4 hours at 37 °C. Absorbance at 450 nm was then measured by a microplate reader, and corresponding proliferation curves were plotted.
Statistics
All statistical analyses were performed using SPSS 20.0. The correlations between expression levels of Sirt3 and patients’ characteristics were tested by chi-squared test. Survival analyses were conducted by Kaplan–Meier method and compared by log-rank test. Multivariate Cox regression analysis was used to identify independent prognostic factors. For the cellular experiments, data were presented as mean ± SD from three independent experiments and compared with Student’s t-test. P<0.05 was considered statistically significant.
Results
Patient information
Our retrospective cohort included 48 females and 66 males, with a median age of 51.0 years. Patients’ clinical characteristics were evaluated by tumor size, WHO grade, and Karnofsky performance score.26 Briefly, 27, 45, and 42 glioma cases were classified as WHO II, WHO III, and WHO IV, respectively. In addition, the surgical strategy was retrieved of all the patients. Forty-eight patients underwent gross total resection, 35 patients underwent subtotal resection, and the other 31 patients underwent partial resection or biopsy. The median overall survival (OS) time was 31.0 months for all the enrolled patients, and the 5-year OS rate is 19.2%. The detailed clinicopathological characteristics of this cohort are described in Table 1.
Sirt3 is upregulated in glioma tissues
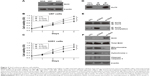
The 114 tissue samples were used for evaluation of Sirt3 protein expression by IHC. According to the distinct staining patterns of Sirt3 (Figure 1A and B), we classified patients into low-expression group and high-expression group. The RT-qPCR results also showed that glioma tissues possessed higher Sirt3 mRNA levels than those of normal brain tissues (Figure 1C, P=0.038). Importantly, by searching the The Cancer Genome Atlas database, we found that low mRNA transcription of Sirt3 indicates a better clinical outcome of glioma patients (Figure 1D, P=0.0097).
Correlation between Sirt3 expression and clinicopathological factors in glioma patients
As shown in Table 1, all 114 glioma patients were divided into high- or low-expression groups based on the median value of the IHC score in tumor tissues. According to chi-squared test, Sirt3 expression was significantly correlated with the WHO grade (P=0.004). Patients with more advanced tumor grade showed higher Sirt3 protein levels.
Sirt3 is a novel biomarker indicating poor prognosis of glioma patients
Because a higher Sirt3 was observed more frequently in glioma patients with advanced WHO grades, we hypothesized that it may help predict patients’ clinical outcomes. We, therefore, analyzed the prognostic factors for the OS in glioma using Kaplan–Meier method (Figure 2, Table 2). A larger tumor size (P=0.027), advanced WHO grades (P=0.012), and noncurative surgical treatment (P<0.001) were shown as unfavorable factors affecting OS. For example, the median OS for patients with WHO grades II, III, and IV was 49.0, 33.0, and 11.0 months, respectively (Table 2). Of note, patients with low Sirt3 expression showed a better prognosis than those with high Sirt3 expression (OS 39.3±2.9 months vs 25.0±3.1 months, P=0.003).
Furthermore, we subjected the significant factors above into a Cox regression multivariate analysis model to explore the independent hazard effect of each factor (Table 3). Although tumor size showed no statistical significance, WHO grade (HR 1.927, 95% CI 1.320–2.814, P=0.001), surgical treatment (HR 1.799, 95% CI 1.164–2.781, P=0.008), and Sirt3 expression (HR 1.602, 95% CI 1.089–2.446, P=0.017) were all independently correlated with overall survival of glioma patients.
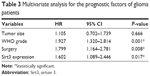  | Table 3 Multivariate analysis for the prognostic factors of glioma patients |
Sirt3 enhances cell viability of glioma cells
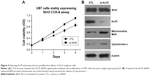
We next tested the functional mechanisms of Sirt3 in glioma. By analyzing its endogenous expression levels in different cell lines, we found that U87 and U251 glioma cells showed significantly higher Sirt3 levels than those in NHA cells (Figure 3A), which is consistent with the RT-qPCR results from clinical tissues samples.
By silencing or overexpressing Sirt3 in both U87 and U251 cells, we further identified that high Sirt3 expression can significantly upregulate the tumor cell viability, as revealed by the CCK-8 assay (Figure 3B and C).
Sirt3 interacts with Ku70 and regulates its acetylation level
There had been evidence that Sirt3 may regulate the function of Ku70 protein in cardiomyocytes during response to oxidative stress.27 Ku70 plays critical roles in controlling the cellular localization of BAX, the mitochondria translocation of which will induce cell apoptosis.28 Moreover, the interaction between Ku70 and BAX was acetylation dependent; once Ku70 is deacetylated, it will release BAX and allow it to transport into mitochondria. Therefore, we performed immunoprecipitation assay to test the existence of Sirt3–Ku70 signaling in U87 glioma cells. As expected, Ku70 was successfully pulled down by immunoprecipitating Sirt3 (Figure 3D). In addition, the acetylation level of Ku70 was negatively correlated with the Sirt3 level (Figure 3E), indicating that Ku70 may be a novel substrate of Sirt3 in glioma cells. Finally, we verified that silencing Sirt3 can significantly increase the BAX level in mitochondria, which subsequently leads to cytochrome c release and caspase 3 cleavage (Figure 3F). In contrast, overexpressing Sirt3 showed completely opposite effects.
Sirt3 may promote tumor progression by targeting Ku70
To further investigate the role of Ku70 in Sirt3 signaling pathway, we next silenced Ku70 in U87 cells that were stably expressing Sirt3. Accordingly, we found that Ku70 siRNA significantly decreased the cell viability (Figure 4A). In addition, the levels of mitochondria BAX and cytosol cytochrome c were enhanced by silencing Ku70, which attenuated the tumor-promoting effects of Sirt3 overexpression (Figure 4B). Thus, we concluded that Sirt3 can promote glioma progression at least partially by Ku70 pathway.
Discussion
Recently, novel strategies on identifying tumor biomarkers are developing rapidly such as the noninvasive platelet screening.29 Another trend is that more and more newly identified biomarkers possess enzymatic functions, especially the phosphorylation, ubiquitination, and acetylation. The protein acetylation mainly occurs on either N terminus or lysine residues. Acetylation is highly dynamic, and its modification level is balanced by acetyltransferases and deacetylases.
A good example on how acetylation regulates tumor progression is the Ku70–BAX signaling pathway. Briefly, the BAX-mediated apoptosis can be suppressed by overexpression of Ku70 in mammalian cells, but enhanced by downregulation of Ku70.30 Following studies demonstrated that Ku70 can directly interact with BAX in cytosol, thus preventing the apoptotic translocation of BAX to mitochondria. It is interesting that the nonacetylation status of Ku70 is a non-negligible prerequisite for the stable interaction between Ku70 and BAX. Under certain apoptotic conditions, the Ku70 is acetylated by BAX, subsequently releases BAX, allows it to transport into mitochondria, and finally induces cell apoptosis. Therefore, it is reasonable that disruption of Ku70–BAX interaction by acetylating Ku70 may lead to tumor cell apoptosis. Indeed, overexpression of CREB-binding protein, a known acetyltransferase toward Ku70, can induce the apoptosis of HeLa cells.31 Consistently, pharmacological inhibiting or silencing Ku70 deacetylase, such as histone deacetylase 6, also successfully triggered BAX-dependent apoptosis of neuroblastoma cells.32 The tumor-related significance of Ku70–BAX interaction had also been revealed in prostate cancer, colon cancer, and osteosarcoma.28,33,34
On the other hand, overexpression of Ku70 upstream deacetylases may exert tumor-promoting functions by inhibiting cell apoptosis. Here we first showed that Sirt3, a member of the Sirt protein family that catalyzes deacetylation, was highly expressed in glioma tissues compared with normal brain tissues. Survival analyses revealed an independent prognostic effect of higher Sirt3 expression on predicating poor prognosis. We then performed cellular experiments to see whether Sirt3 can directly regulate glioma cell viability. As expected, overexpression of Sirt3 enhanced cell growth, while silencing Sirt3 via siRNA significantly promoted cell apoptosis. Furthermore, our data demonstrated the involvement of Ku70 deacetylation in Sirt3 signaling pathways, which was consistent with its enzymatic relationship in cardiomyocytes.27 Consistently, a recent study reported that treatment by petunidin-3-O-glucoside (Pt3glc) displayed potent antiproliferative effects and also decreased the Sirt3 protein level.35 Further investigation is needed to determine whether Pt3glc functions by directly targeting Sirt3. Therefore, targeting the expression of Sirt3 may be another novel therapeutic direction for aging-associated tumors.36 Finally, we verified that Sirt3 overexpression inhibited glioma cell apoptosis by stabilizing Ku70–BAX interaction in cytosol.
Our study therefore provided evidence of the potential function of Sirt3 in glioma progression. Since several high throughput strategies are being used to screen Sirt3 inhibitors, it is highly likely that Sirt3 will be a novel therapeutic drug target in the near future.37–39 Moreover, several groups are focusing on developing drugs targeting the Ku70–BAX interaction,33,40 which may be combined with Sirt3 inhibitors in treating malignancies. A similar example is the interaction between Sirt3 and superoxide dismutase 2 (SOD2) protein. Cheng et al reported that inhibiting the Sirt3 expression or disrupting Sirt3–SOD2 interaction may help attenuate the drug resistance of linalool, a specific drug targeting SOD2, on glioma cells.41
Conclusion
In summary, our study revealed the expression patterns of Sirt3 in glioma patients for the first time and demonstrated its correlations with the disease stages and prognosis. Identification of Sirt3 as a novel biomarker for glioma and revealing its functional mechanisms would be invaluable for treatment improvement.
Disclosure
The authors report no conflicts of interest in this work.
References
Deangelis LM. Brain tumors. N Engl J Med. 2001;344(2):114–123. | ||
Gao J, Ciriello G, Sander C, Schultz N. Collection, integration and analysis of cancer genomic profiles: from data to insight. Curr Opin Genet Dev. 2014;24:92–98. | ||
Liu L, Li W, Geng S, et al. Slit2 and Robo1 expression as biomarkers for assessing prognosis in brain glioma patients. Surg Oncol. 2016;25(4):405–410. | ||
Siegel RL, Miller KD, Jemal A. Cancer Statistics. CA Cancer J Clin. 2018;68(1):7–30. | ||
Li X, Zheng J, Diao H, Liu Y. RUNX3 is down-regulated in glioma by Myc-regulated miR-4295. J Cell Mol Med. 2016;20(3):518–525. | ||
Osborne B, Bentley NL, Montgomery MK, Turner N. The role of mitochondrial sirtuins in health and disease. Free Radic Biol Med. 2016;100:164–174. | ||
Winnik S, Auwerx J, Sinclair DA, Matter CM. Protective effects of sirtuins in cardiovascular diseases: from bench to bedside. Eur Heart J. 2015;36(48):3404–3412. | ||
Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287(17):14078–14086. | ||
Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21(8):920–928. | ||
Koentges C, Pfeil K, Meyer-Steenbuck M, et al. Preserved recovery of cardiac function following ischemia-reperfusion in mice lacking SIRT3. Can J Physiol Pharmacol. 2016;94(1):72–80. | ||
Torrens-Mas M, Oliver J, Roca P, Sastre-Serra J. SIRT3: oncogene and tumor suppressor in cancer. Cancers. 2017;9(12):90. | ||
Desouki MM, Doubinskaia I, Gius D, Abdulkadir SA. Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer. Hum Pathol. 2014;45(5):1071–1077. | ||
Liu Y, Liu YL, Cheng W, Yin XM, Jiang B. The expression of SIRT3 in primary hepatocellular carcinoma and the mechanism of its tumor suppressing effects. Eur Rev Med Pharmacol Sci. 2017;21(5):978–998. | ||
Yu W, Denu RA, Krautkramer KA, et al. Loss of SIRT3 provides growth advantage for B cell malignancies. J Biol Chem. 2016;291(7):3268–3279. | ||
Liu C, Huang Z, Jiang H, Shi F. The sirtuin 3 expression profile is associated with pathological and clinical outcomes in colon cancer patients. Biomed Res Int. 2014;2014:1–9. | ||
Cui Y, Qin L, Wu J, et al. SIRT3 enhances glycolysis and proliferation in SIRT3-expressing gastric cancer cells. PLoS One. 2015;10(6):e0129834. | ||
George J, Nihal M, Singh CK, Zhong W, Liu X, Ahmad N. Pro-proliferative function of mitochondrial sirtuin deacetylase SIRT3 in human melanoma. J Invest Dermatol. 2016;136(4):809–818. | ||
Choi J, Koh E, Lee YS, et al. Mitochondrial Sirt3 supports cell proliferation by regulating glutamine-dependent oxidation in renal cell carcinoma. Biochem Biophys Res Commun. 2016;474(3):547–553. | ||
Zhang Q, Fan H, Zou Q, et al. TEAD4 exerts pro-metastatic effects and is negatively regulated by miR6839-3p in lung adenocarcinoma progression. J Cell Mol Med. 2018;22(7):3560–3571. | ||
Liu H, Xu Y, Zhang Q, et al. Correlations between TBL1XR1 and recurrence of colorectal cancer. Sci Rep. 2017;7:44275. | ||
Xu Y, Yang X, Li Z, et al. Sprouty2 correlates with favorable prognosis of gastric adenocarcinoma via suppressing FGFR2-induced ERK phosphorylation and cancer progression. Oncotarget. 2017;8(3):4888. | ||
Tan W, Pan M, Liu H, Tian H, Ye Q, Liu H. Ergosterol peroxide inhibits ovarian cancer cell growth through multiple pathways. Onco Targets Ther. 2017;10:3467–3474. | ||
Liu H, Zhang Q, Li K, et al. Prognostic significance of USP33 in advanced colorectal cancer patients: new insights into β-arrestin-dependent ERK signaling. Oncotarget. 2016;7(49):81223–81240. | ||
Fly R, Lloyd J, Krueger S, Fernie A, van der Merwe MJ. Improvements to define mitochondrial metabolomics using nonaqueous fractionation. Methods Mol Biol. 2015;1305:197–210. | ||
Hou S, du P, Wang P, Wang C, Liu P, Liu H. Significance of MNK1 in prognostic prediction and chemotherapy development of epithelial ovarian cancer. Clin Transl Oncol. 2017;19(9):1107–1116. | ||
Johnson MJ, Bland JM, Davidson PM, et al. The relationship between two performance scales: New York Heart Association Classification and Karnofsky Performance Status Scale. J Pain Symptom Manage. 2014;47(3):652–658. | ||
Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28(20):6384–6401. | ||
Trougakos IP, Lourda M, Antonelou MH, et al. Intracellular clusterin inhibits mitochondrial apoptosis by suppressing p53-activating stress signals and stabilizing the cytosolic Ku70-Bax protein complex. Clin Cancer Res. 2009;15(1):48–59. | ||
Zhang Q, Liu H, Zhu Q, et al. Patterns and functional implications of platelets upon tumor “education.” Int J Biochem Cell Biol. 2017;90:68–80. | ||
Sawada M, Sun W, Hayes P, Leskov K, Boothman DA, Matsuyama S. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nat Cell Biol. 2003;5(4):320–329. | ||
Cohen HY, Lavu S, Bitterman KJ, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13(5):627–638. | ||
Subramanian C, Jarzembowski JA, Opipari AW, Castle VP, Kwok RP. HDAC6 deacetylates Ku70 and regulates Ku70-Bax binding in neuroblastoma. Neoplasia. 2011;13(8):726–734. | ||
Shukla S, Fu P, Gupta S. Apigenin induces apoptosis by targeting inhibitor of apoptosis proteins and Ku70-Bax interaction in prostate cancer. Apoptosis. 2014;19(5):883–894. | ||
Pucci S, Mazzarelli P, Paola M, Sesti F, et al. Interleukin-6 affects cell death escaping mechanisms acting on Bax-Ku70-Clusterin interactions in human colon cancer progression. Cell Cycle. 2009;8(3):473–481. | ||
Wang G, Fu XL, Wang JJ, Guan R, Sun Y, To ST. Inhibition of glycolytic metabolism in glioblastoma cells by Pt3glc combinated with PI3K inhibitor via SIRT3-mediated mitochondrial and PI3K/Akt-MAPK pathway. J Cell Physiol. Epub 16 Jan 2018. | ||
Son MJ, Ryu JS, Kim JY, et al. Upregulation of mitochondrial NAD+ levels impairs the clonogenicity of SSEA1+ glioblastoma tumor-initiating cells. Exp Mol Med. 2017;49(6):e344. | ||
Salo HS, Laitinen T, Poso A, Jarho E, Lahtela-Kakkonen M. Identification of novel SIRT3 inhibitor scaffolds by virtual screening. Bioorg Med Chem Lett. 2013;23(10):2990–2995. | ||
Disch JS, Evindar G, Chiu CH, et al. Discovery of thieno[3,2-d]pyrimidine-6-carboxamides as potent inhibitors of SIRT1, SIRT2, and SIRT3. J Med Chem. 2013;56(9):3666–3679. | ||
Chen Y, Fu LL, Wen X, et al. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 2014;5(2):e1047. | ||
Sawada M, Hayes P, Matsuyama S. Cytoprotective membrane-permeable peptides designed from the Bax-binding domain of Ku70. Nat Cell Biol. 2003;5(4):352–357. | ||
Cheng Y, Dai C, Zhang J. SIRT3-SOD2-ROS pathway is involved in linalool-induced glioma cell apoptotic death. Acta Biochim Pol. 2017;64(2):343–350. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.