Back to Journals » Infection and Drug Resistance » Volume 11
Simultaneous three Enterobacteriaceae with different bla NDM-1-encoding plasmids in a patient transferred from mainland China to Taiwan
Authors Lai CC , Chen CC , Lu YC , Chen HJ, Su BA, Weng TC, Chiu YH, Toh HS , Zhang CC, Chiang SR, Chuang YC, Tang HJ
Received 3 July 2018
Accepted for publication 3 November 2018
Published 7 December 2018 Volume 2018:11 Pages 2555—2560
DOI https://doi.org/10.2147/IDR.S179024
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Chih-Cheng Lai,1 Chi-Chung Chen,2,3 Ying-Chen Lu,3 Hung-Jui Chen,4 Bo-An Su,4 Tzu-Chieh Weng,4 Yu-Hsin Chiu,5 Han-Siong Toh,4 Chun-Cheng Zhang,4 Shyh-Ren Chiang,4 Yin-Ching Chuang,2,5 Hung-Jen Tang2,4,6
1Department of Intensive Care Medicine, Chi Mei Medical Center, Liouying, Tainan, Taiwan; 2Department of Medical Research, Chi Mei Medical Center, Tainan, Taiwan; 3Department of Food Science, National Chiayi University, Chiayi, Taiwan; 4Department of Medicine, Chi Mei Medical Center, Tainan, Taiwan; 5Department of Internal Medicine, Chi Mei Medical Center, Liouying, Tainan, Taiwan; 6Department of Health and Nutrition, Chia Nan University of Pharmacy and Science, Tainan, Taiwan
Abstract: New-Delhi metallo-β-lactamase1 (NDM-1) Enterobacteriaceae are increasing worldwide. Herein, we describe a single patient who carried three unusual NDM-1 carbapenem-resistant Enterobacteriaceae – Enterobacter cloacae (E. cloacae) yielded from a urine specimen and Klebsiella pneumoniae (K. pneumoniae) and Escherichia coli (E. coli) from stool specimens. For E. cloacae, its bla NDM-1-encoding plasmid was pKP04NDM with a size of ~54 kb replicons with an IncN backbone. For K. pneumoniae, its bla NDM-1-encoding plasmid was pNDM-BTR with a size of ~59.6 kb and belonged to IncN. For E. coli, its main bla NDM-1-encoding plasmid was pIMP-HK1500, and the NDM-1 gene was obtained from a part of pNDM-BTR (8439 bp). These three clinical strains are reported for the first time and are assumed to be imported from mainland China to Taiwan. The three different plasmids were never reported in K. pneumoniae, E. coli, and Citrobacter spp before. Owing to their associated multidrug resistance, appropriate measures of periodic, targeted surveillance, and development of new antimicrobial agents are urgently needed.
Keywords: carbapenem-resistant Enterobacteriaceae, NDM-1, K. pneumoniae, E. coli, Citrobacter, plasmid
Introduction
Carbapenem-resistant Enterobacteriaceae are increasing worldwide and have become a severe public health threat.1 Several mechanisms of carbapenem resistance, including the production of extended spectrum β-lactamase (ESBL) and AmpC enzymes, and secretions of carbapenemase, have been reported. In terms of carbapenemases, Klebsiella pneumoniae carbapenemase (KPC) and New-Delhi metallo-β-lactamase1 (NDM-1) are the most notorious because they are associated with high-level carbapenem resistance and can spread between different species of Enterobacteriaceae.2 This critical condition was also noted in Taiwan,3,4 and the National Task Force of the Carbapenem Resistance Monitoring Program was implemented. Through this program, we first detected a single patient who carried three unusual NDM-1 producing carbapenem-resistant Enterobacteriaceae.
Methods
Bacterial strains
Three strains, carbapenem-resistant Enterobacter cloacae (E. cloacae) CRE961, K. pneumoniae CRE967, and Escherichia. coli (E. coli) CRE968, were isolated from a 30-year-old, previously healthy woman who was transferred from mainland China and hospitalized at Chi-Mei Medical Center in Taiwan for spontaneous brainstem hemorrhage. Because of her vegetative status, she received a tracheostomy and prolonged mechanical ventilation in mainland China. The patient was initially hospitalized in mainland China for one month and then transferred to Taiwan for long-term care. E. cloacae CRE961 was isolated from the first urine specimen in Taiwan and persisted during the whole course. One month after admission in Taiwan, the patient had diarrhea, and we sent the stool specimen for an initial examination. E. cloacae CRE961 was yielded from the urine specimen on April 24 and May 3 in 2017 and K. pneumoniae CRE967 and E. coli CRE968 were yielded from stool specimens on May 25. These bacterial species were confirmed by a VITEK 2 automated system (bioMe ´rieux, Marcy l’Etoile, France) with VITEK®2 GN ID card. These isolates were stored at –80°C in Protect Bacterial Preservers (Technical Service Consultants Limited, Heywood, UK) before investigation. The patient’s written informed consent was obtained; however, no ethical approval was required for this case from the Chi Mei Medical Center as this study was based on retrospective design with routine laboratory work.
Antimicrobial susceptibility testing and minimum inhibitory concentration (MIC) measurement
Standard powders of amikacin, ciprofloxacin, doxycycline, ertapenem, gentamicin, imipenem (U.S. Pharmacopeia, Rockville, MD, USA), ampicillin, cephalothin, cefuroxime, ceftriaxone, ceftazidime, colistin sulfate, doripenem, meropenem, trimethoprim/sulfamethoxazole (Sigma-Aldrich, St. Louis, MO, USA), fosfomycin (Ercros, Barcelona, Spain), and tigecycline (Pfizer, New York, NY, USA) were used for antimicrobial susceptibility tests. MIC determinations and susceptibility interpretation criteria followed the Clinical Laboratory and Standard Institute (CLSI) and Federal Drug Administration standards.5,6 MICs of the drugs, except tigecycline and colistin, were measured by agar dilution in Mueller–Hinton agar (Oxoid, Basingstoke, UK) according to CLSI recommendations.5 For fosfomycin susceptibility, glucose-6-phosphate (25 mg/mL) was added to the agar plate. Tigecycline and colistin MICs were determined by broth microdilutions in freshly prepared cation-adjusted Mueller–Hinton broth.8 E. coli ATCC 25922 was used as the control strain.
Multilocus sequence typing (MLST)
MLST was performed with seven housekeeping genes (E. cloacae CRE961, including dnaA, fusA, gyrB, leuS, pyrG, rplB, and rpoB; K. pneumoniae CRE967, including gapA, infB, mdh, pgi, phoE, rpoB, and tonB, and E. coli CRE968, including adk, fumC, gyrB, icd, mdh, purA, and recA) as previously described.7–9 The allele sequences and STs were verified at http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html.
DNA manipulation and PCR amplification
Genomic DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Plasmid DNA was also extracted by a QIAprep Spin Miniprep kit (Qiagen). PCR amplifications were performed using specific primers as previously described.7
PCR detection and sequencing of antibiotic resistance genes
PCR was used to amplify the ESBL genes (blaTEM, blaSHV, blaCTX-M), ampC genes (blaDHA-1 and blaCMY-2) and to screen the representative carbapenemase gene (blaKPC-2, blaNDM) using specific primers as previously published.10–13 Amplicons of β-lactamase genes were purified with PCR clean-up kits (Roche Diagnostics GmbH, Penzberg, Germany) and were sequenced on an ABI PRISM 3730 sequencer analyzer (Applied Biosystems, Foster City, CA, USA).7
S1-nuclease pulsed-field gel electrophoresis (PFGE)
Plasmid DNA was extracted from bacteria with the Qiagen Midi Kit (Qiagen). Plasmid sizing was performed using S1-nuclease (Promega, Madison, WI, USA) digested plasmid DNA, and then separated by PFGE using a CHEF mapper system (Bio-Rad, Berkeley, CA, USA) as previously described.14
Southern Blot of NDM-1
Southern Blotting was performed using semi-dry transfer system (Bio-Rad) and NDM-containing plasmids were identified by hybridization with Dig-labeled blaNDM-specific probe generated by the PCR DIG Probe Synthesis Kit, and Detection Starter Kit II (Roche Applied Sciences, Mannheim, Germany) as previously reported.14
Plasmid sequencing
Bacterial pellets in centrifuge tubes were resuspended in buffer. Cell wall was removed by enzymatic digestion in the presence of RNase. Cell lysis and chromosome removal was achieved by alkaline lysis, followed by acid aggregation and centrifugation. DNA in the supernatant was extracted using organic solvent and recovered by ethanol precipitation. Concentration of samples was determined by fluorescence quantification. Purified DNA was analyzed by electrophoresis in 0.4% agarose gel matrix. The Illumina MiSeq System (Illumina, San Diego, CA, USA) was used for plasmid sequencing. The derived reads were assembled using the CLC Genomics Workbench 5.51 (CLC bio, Aarhus, Denmark).15
Results
Antibiotic susceptibility
The MIC values of the antibiotics against three CRE strains are shown in Table 1. All of them were susceptible to tigecycline and colistin but resistant to all carbapenems, including imipenem, meropenem, ertapenem, and doripenem. In addition, these three strains were resistant to most of other antibiotics.
  | Table 1 Minimal inhibitory concentrations of various antibiotics against three Enterobacteriaceae |
Molecular characteristics
Based on the MLST, these three strains belonged to different ST types, E. cloacae (ST932), K. pneumoniae (ST656), and E. coli (ST131), which were detected by different housekeeping genes. Plasmid DNA was converted to a linear form by S1-nuclease on PFGE and agarose gel showing the S1-nuclease PFGE-based sizing of plasmids for 3 isolates (Figure 1A, arrows). Figure 1B shows the corresponding gel after Southern Blotting, and the plasmids with NDM-1 were detected by Southern Blot with a specific probe.
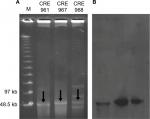  | Figure 1 (A) S1-nuclease pulsed-field gel electrophoresis profiles and (B) Southern Blot of three Enterobacteriaceae carrying the NDM-1 plasmid. |
Antibiotic-resistant gene
All three isolates were positive for blaESBL, and the gene encoding CTX-M-55 was detected for K. pneumoniae CRE967, and E. coli CRE968. The genes encoding SHV-12 and SHV-31 were detected for E. cloacae CRE961, and K. pneumoniae CRE967, respectively. Only one strain, K. pneumoniae CRE967, was positive for blaampc, which is where the gene encoding DHA-1 was found. In addition, the gene encoding TEM was found for K. pneumoniae CRE967, and E. coli CRE968.
Sequencing analyses of the carbapenem-resistant plasmids
For E. cloacae CRE961, its blaNDM-1-encoding plasmid was pKP04NDM (GenBank accession no. KU314941.1), with a size of ~54 kb, and an IncN backbone (Figure 2A). For K. pneumoniae CRE967, its blaNDM-1-encoding plasmid was pNDM-BTR (GenBank accession no. KF534788.2) with a size of ~59.6 kb and belonging to IncN (Figure 2B). For E. coli CRE968, its main blaNDM-1-encoding plasmid was pIMP-HK1500 (GenBank accession no. KT989599.1), and the NDM-1 gene was obtained from a part of pNDM-BTR (8439 bp). Its size was 60.7 kb and belonged to IncX3 (Figure 2C).
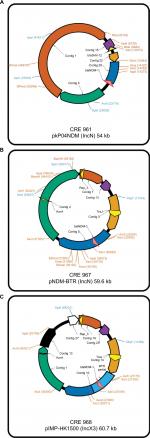  | Figure 2 Schematic diagrams of different plasmids (A) Enterobacter cloacae CRE961 (pKP04NDM), (B) Klebsiella pneumoniae CRE967 (pNDM-BTR), and (C) Escherichia coli CRE968 (pIMP-HK1500). |
Discussion
Since NDM-1 was first identified in a K. pneumoniae isolate in India in 2009,16 more and more carbapenem-resistant Enterobacteriaceae have been found to carry NDM-1.17,18 In this report, we present a rare case of three different NDM-1 carbapenem-resistant Enterobacteriaceae that were isolated from the clinical specimens of one patient during a 1-month hospitalization. It is difficult to identify the sources of the three different unusual NDM-1 Enterobacteriaceae of this patient. Clinically, these three NDM-1 Enterobacteriaceae were considered as colonization strains. During the hospitalization in Taiwan, the patient ever received the following antibiotics – levofloxacin, ceftriaxone, minocycline, and amoxicillin/clavulanate. According to the clinical history, it was thought that all of three NDM-1 Enterobacteriaceae were carried by this patient from mainland China to Taiwan.
IncX3 and IncN were two major plasmids found in three NDM-1 Enterobacteriaceae. IncX plasmids are thought to be narrow-host-range plasmids of Enterobacteriaceae and are low in prevalence. Some studies from China have shown that the most common plasmid Inc type to harbor blaNDM was IncX3, indicating that the main manner mediating the transfer of blaNDM may be the spread of IncX3 plasmids in China.19 Previous reports also indicated that the IncN plasmid has been shown to encode clinically important resistance determinants among E. coli and also in K. pneumoniae isolates, such as blaCTX-M and blaNDM-1.20 Highly efficient transmission of the IncN and IncX plasmids appeared to account for the diversity and worldwide spread of blaNDM-1-carrying Enterobacteriaceae just as the results found in our three NDM-1 Enterobacteriaceae.
E. cloacae CRE961 belongs to ST932, which has never been identified in Taiwan or the whole world, and its blaNDM-1-encoding plasmid is pKP04NDM. The genetic environment surrounding blaNDM-1 was identical to that found in various blaNDM-1-carrying plasmids in Enterobacteriaceae in mainland China, including pKP04NDM (KU314941), pNDM-HN380 (JX104760), pKPN5047 (KC311431), and pNDM-SX04 (KC876051) in K. pneumoniae.21 In other words, plasmid pKP04NDM was generally found in K. pneumoniae, which is the first reported E. cloacae ST 932 that carried such a plasmid with the NDM-1 gene.
K. pneumoniae CRE967, which belongs to ST656, has been found in mainland China22,23and the Philippines,23 but not Taiwan. Its blaNDM-1-encoding plasmid pNDM-BTR has been found in E. coli strains in mainland China.24 Generally, plasmid pNDM-BTR is found in E. coli ST131. However, this is the first report of a K. pneumoniae ST656 that carried such an NDM-1 plasmid.
E. coli CRE968 belongs to ST131 and has been identified in Thailand25 and mainland China.26 Its main blaNDM-1-encoding plasmid is pIMP-HK1500, and the NDM-1 gene is obtained from a part of pNDM-BTR (8439 bp). Insertion sequences were found on both sides of NDM-1 gene in pNDM-BTR. Therefore, we speculate the insertion sequence was inserted into pIMP-HK1500 plasmid direct or indirectly.27 Plasmid pIMP-HK1500 is generally found in Citrobacter spp,28 but a plasmid that carried the NDM-1 gene has never been reported before among E. coli ST131 due to most of them carrying the NDM-1-carrying plasmid pNDM-BTR. In addition to NDM-1, co-carriage of different ESBL and AmpC genes in combination were noted among one of the NDM-1 isolates, K. pneumoniae CRE967, in the present report.
Initially, we thought that the three different blaNDM-1 isolates in one patient may have been caused by plasmid transformation or conjugation among different bacteria. However, our analysis of different plasmids revealed that they not only harbor the NDM-1 gene but also different characteristics among the plasmids. In addition, according to the plasmid analysis results, we can also find the possible plasmid transformation among different Enterobacteriaceae, which seems to be more severe in mainland China; further infection control intervention should be implemented in the future.
In this report, all three NDM-1 carbapenem-resistant Enterobacteriaceae were resistant to all the tested carbapenems, including imipenem, meropenem, and doripenem, but remained susceptible to tigecycline and colistin. This is consistent with previous reports29 regarding NDM-1 in Taiwan. Although the number is limited, the result suggests that colistin or tigecycline may be the drug of choice for these pathogens in Taiwan.
In conclusion, we identified the first patient with three different NDM-1 carbapenem-resistant Enterobacteriaceae isolated during a single 1-month hospitalization. Most importantly, E. cloacae ST-932 with a blaNDM-1-encoding plasmid pKP04NDM, K. pneumoniae ST-656 with a blaNDM-1-encoding plasmid pNDM-BTR, and E. coli ST-131 with a main plasmid pIMP-HK1500 inserted with the NDM-1 gene are reported for the first time. Owing to their associated multidrug resistance, appropriate measures of periodic, targeted surveillance, and development of new antimicrobial agents are urgently needed not only in Taiwan but around the world.
Acknowledgments
This study was supported by a research grant (no. CMFHT10501, CMNCKU10509) from the Chi-Mei Medical Center Research Foundation, and MOST 105-2314-B-384-007-MY3 from Ministry of science and technology.
Disclosure
The authors report no conflicts of interest in this work.
References
Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(Suppl 1):S28–S36. | ||
Poirel L, Pitout JD, Nordmann P. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2007;2(5):501–512. | ||
Wang JT, Wu UI, Lauderdale TL, et al. Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PLoS One. 2015;10(3):e0121668. | ||
Tang HJ, Hsieh CF, Chang PC, et al. Clinical significance of community- and healthcare-acquired carbapenem-resistant Enterobacteriaceae isolates. PLoS One. 2016;11(3):e0151897. | ||
Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: 16th informational supplement. Document M100-S22. Wayne, PA: CLSI; 2012. | ||
Brown SD, Traczewski MM. Comparative in vitro antimicrobial activity of tigecycline, a new glycylcycline compound, in freshly prepared medium and quality control. J Clin Microbiol. 2007;45(7):2173–2179. | ||
Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. | ||
Ma L, Siu LK, Lin JC, et al. Updated molecular epidemiology of carbapenem-non-susceptible Escherichia coli in Taiwan: first identification of KPC-2 or NDM-1-producing E. coli in Taiwan. BMC Infect Dis. 2013;13:599. | ||
Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One. 2013;8(6):e66358. | ||
Tang HJ, Ku YH, Lee MF, Chuang YC, Yu WL. In vitro activity of imipenem and colistin against a carbapenem-resistant Klebsiella pneumoniae isolate coproducing SHV-31, CMY-2, and DHA-1. Biomed Res Int. 2015;2015:568079. | ||
Eckert C, Gautier V, Saladin-Allard M, et al. Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob Agents Chemother. 2004;48(4):1249–1255. | ||
Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. | ||
Poirel L, Revathi G, Bernabeu S, Nordmann P. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother. 2011;55(2):934–936. | ||
Mshana SE, Hain T, Domann E, et al. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis. 2013;13:466. | ||
Scott D, Ely B. Comparison of genome sequencing technology and assembly methods for the analysis of a GC-rich bacterial genome. Curr Microbiol. 2015;70(3):338–344. | ||
Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. | ||
Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013;62(Pt 4):499–513. | ||
Khan AU, Maryam L, Zarrilli R. Structure, genetics and worldwide spread of New Delhi Metallo-beta-lactamase (NDM): a threat to public health. BMC Microbiol. 2017;17(1):101. | ||
An J, Guo L, Zhou L, et al. NDM-producing Enterobacteriaceae in a Chinese hospital, 2014-2015: identification of NDM-producing Citrobacter werkmanii and acquisition of blaNDM-1-carrying plasmid in vivo in a clinical Escherichia coli isolate. J Med Microbiol. 2016;65(11):1253–1259. | ||
Du J, Li B, Cao J, et al. Molecular characterization and epidemiologic study of NDM-1-producing extensively drug-resistant Escherichia coli. Microb Drug Resist. 2017;23(3):272–279. | ||
Zhong LL, Zhang YF, Doi Y, et al. Coproduction of MCR-1 and NDM-1 by colistin-resistant Escherichia coli isolated from a healthy individual. Antimicrob Agents Chemother. 2017;61(1):e01962–16. | ||
Liu H, Wilksch J, Li B, et al. Emergence of ST39 and ST656 extensively drug-resistant Klebsiella pneumoniae isolates in Wenzhou, China. Indian J Med Microbiol. 2017;35(1):145–146. | ||
Chou A, Roa M, Evangelista MA, et al. Emergence of Klebsiella pneumoniae ST273 Carrying blaNDM-7 and ST656 Carrying blaNDM-1 in Manila, Philippines. Microb Drug Resist. 2016;22(7):585–588. | ||
Zhao Y, Wang L, Zhang Z, et al. Structural genomics of pNDM-BTR harboring In191 and Tn6360, and other bla NDM-carrying IncN1 plasmids. Future Microbiol. 2017;12:1271–1281. | ||
Netikul T, Sidjabat HE, Paterson DL, et al. Characterization of an IncN2-type blaNDM-1-carrying plasmid in Escherichia coli ST131 and Klebsiella pneumoniae ST11 and ST15 isolates in Thailand. J Antimicrob Chemother. 2014;69(11):3161–3163. | ||
Wang X, Chen G, Wu X, et al. Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the bla NDM-1 element and clonal spread of progenitor resistant strains. Front Microbiol. 2015;6:595. | ||
Campos JC, da Silva MJ, dos Santos PR, et al. Characterization of Tn3000, a transposon responsible for blaNDM-1 dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob Agents Chemother. 2015;59(12):7387–7395. | ||
Wang Y, Lo WU, Lai RW, et al. IncN ST7 epidemic plasmid carrying blaIMP-4 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. J Antimicrob Chemother. 2017;72(1):99–103. | ||
Chen CJ, Wu TL, Lu PL, et al. Closely related NDM-1-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. PLoS One. 2014;9(8):e104899. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
